1P7D
 
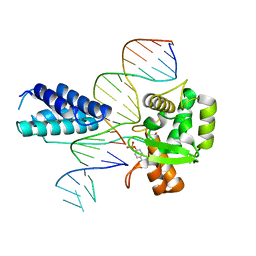 | | Crystal structure of the Lambda Integrase (residues 75-356) bound to DNA | | Descriptor: | 26-MER, 5'-D(*CP*AP*AP*TP*GP*CP*CP*AP*AP*CP*TP*TP*T)-3', Integrase | | Authors: | Aihara, H, Kwon, H.J, Nunes-Duby, S.E, Landy, A, Ellenberger, T. | | Deposit date: | 2003-05-01 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Conformational Switch Controls the DNA Cleavage Activity of Lambda Integrase
Mol.Cell, 12, 2003
|
|
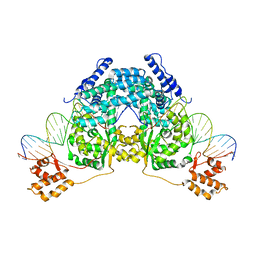
2V6E
 
 | |
1AA3
 
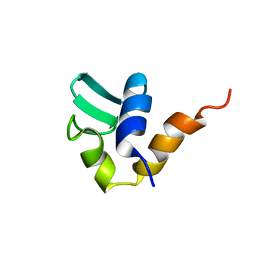 | | C-TERMINAL DOMAIN OF THE E. COLI RECA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RECA | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Terada, T, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing.
J.Mol.Biol., 274, 1997
|
|
1B22
 
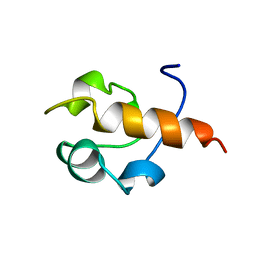 | | RAD51 (N-TERMINAL DOMAIN) | | Descriptor: | DNA REPAIR PROTEIN RAD51 | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
6DFY
 
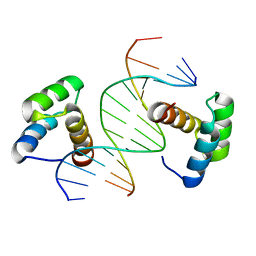 | | Remodeled crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*A)-3'), Double homeobox protein 4 | | Authors: | Aihara, H, Shi, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Comment on structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
4YJ0
 
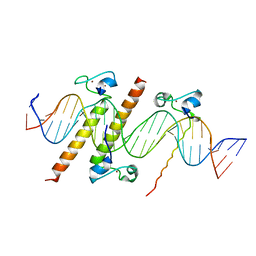 | | Crystal structure of the DM domain of human DMRT1 bound to 25mer target DNA | | Descriptor: | DNA (25-MER), Doublesex- and mab-3-related transcription factor 1, ZINC ION | | Authors: | Murphy, M.W, Lee, J.K, Rojo, S, Gearhart, M.D, Kurahashi, K, Banerjee, S, Loeuille, G, Bashamboo, A, McElreavey, K, Zarkower, D, Aihara, H, Bardwell, V.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | An ancient protein-DNA interaction underlying metazoan sex determination.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8SBM
 
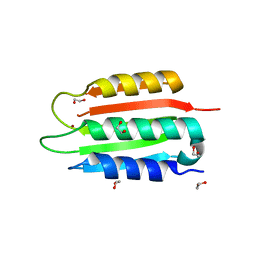 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | Descriptor: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | Authors: | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | Deposit date: | 2023-04-03 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
6XC0
 
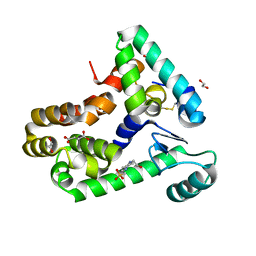 | | Crystal structure of bacteriophage T4 spackle and lysozyme in monoclinic form | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-06-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
4F41
 
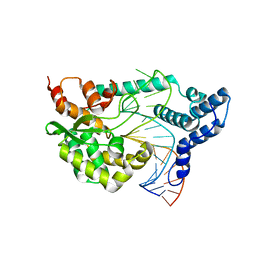 | |
7R9C
 
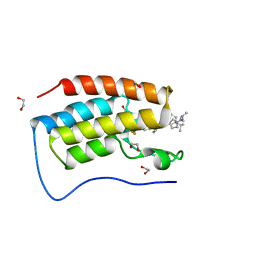 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
4FPV
 
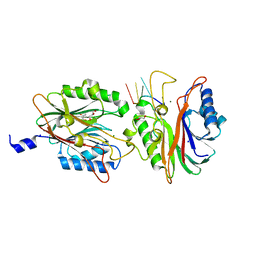 | | Crystal structure of D. rerio TDP2 complexed with single strand DNA product | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FW2
 
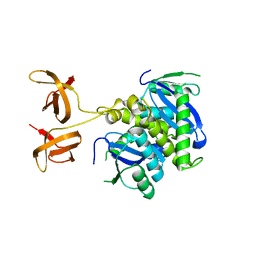 | |
4NQ3
 
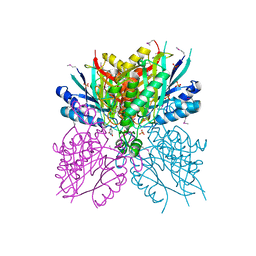 | | Crystal structure of cyanuic acid hydrolase from A. caulinodans | | Descriptor: | BARBITURIC ACID, Cyanuric acid amidohydrolase, MAGNESIUM ION, ... | | Authors: | Cho, S, Shi, K, Aihara, H. | | Deposit date: | 2013-11-23 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Cyanuric acid hydrolase from Azorhizobium caulinodans ORS 571: crystal structure and insights into a new class of Ser-Lys dyad proteins.
Plos One, 9, 2014
|
|
6U81
 
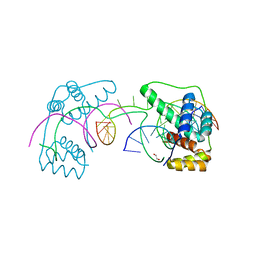 | | Crystal Structure of the Double Homeodomain of DUX4 in Complex with a DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*CP*CP*CP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2019-09-04 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA aptamers against the DUX4 protein reveal novel therapeutic implications for FSHD.
Faseb J., 34, 2020
|
|
8VJY
 
 | |
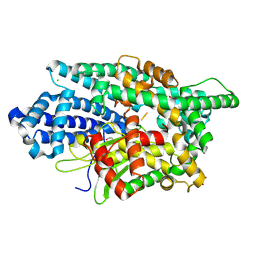
8VJU
 
 | |
8VJV
 
 | |
8VJW
 
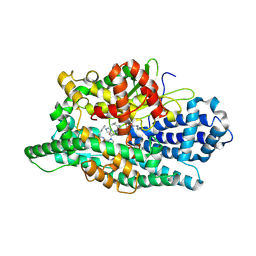 | |
8VA0
 
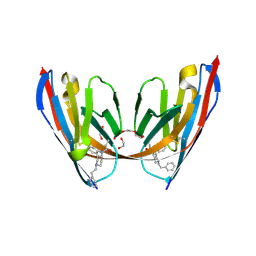 | | X-ray crystal structure of JGFN4 N76D complexed with fentanyl in dimer form | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Shi, K, Moller, N, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8V9W
 
 | | X-ray crystal structure of JGFN4 complexed with fentanyl | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Moller, N, Shi, K, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
9BZG
 
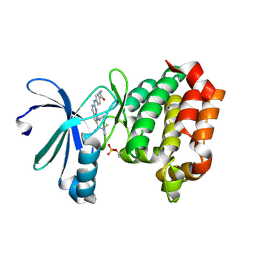 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-[4-(methylcarbamoyl)anilino]-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
9BZL
 
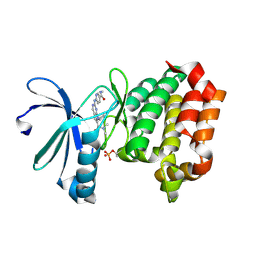 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-{[5-(methylcarbamoyl)pyridin-2-yl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
6U82
 
 | |
7TV2
 
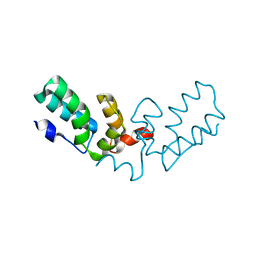 | | X-ray crystal structure of HIV-2 CA protein CTD | | Descriptor: | Capsid protein p24 | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2022-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation.
J.Mol.Biol., 435, 2023
|
|
8SPH
 
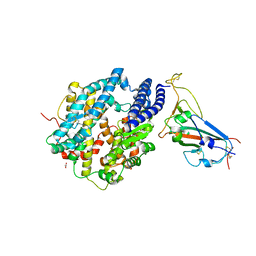 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
