3ID0
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with 3-Fluoro-1-(2-hydroxy-2,2-bisphosphono-ethyl)pyridinium | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-19 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
6NCI
 
 | | Crystal structure of CDP-Chase: Vector data collection | | Descriptor: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
3IBA
 
 | | Crystal structure of the complex of Trypanosoma cruzi farnesyl diphosphate synthase with zoledronate, IPP and Mg2+ | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
3ICK
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with minodronate and isopentenyl disphosphate | | Descriptor: | (1-HYDROXY-2-IMIDAZO[1,2-A]PYRIDIN-3-YLETHANE-1,1-DIYL)BIS(PHOSPHONIC ACID), 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ACETIC ACID, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-17 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
6NCH
 
 | | Crystal structure of CDP-Chase: Raster data collection | | Descriptor: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
3ICN
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with isopentenyl pyrophosphate and 3-Fluoro-1-(2-hydroxy-2,2-bis-phosphono-ethyl)-pyridinium | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-17 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
3ICM
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with isopentenyl pyrophosphate, Mg2+ and 1-(2-Hydroxy-2,2-bis-phosphono-ethyl)-3-phenyl-pyridinium | | Descriptor: | 1-(2-hydroxy-2,2-diphosphonoethyl)-3-phenylpyridinium, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-17 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
6NCK
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Miller, M.S, Maheshwari, S, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
7R63
 
 | |
7UXW
 
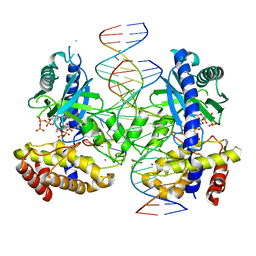 | | Structure of ATP and GTP bind to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UYQ
 
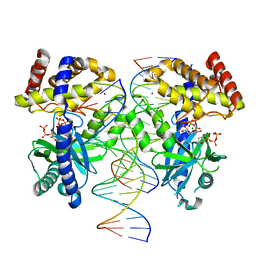 | | Structure of GTP binds to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UYZ
 
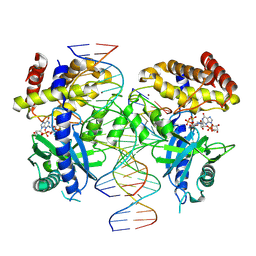 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UZR
 
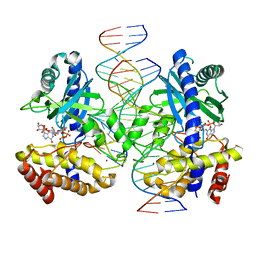 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, MANGANESE (II) ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
6BUU
 
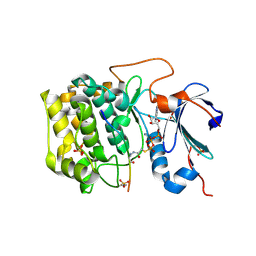 | | Crystal structure of AKT1 (aa 144-480) with a bisubstrate | | Descriptor: | GLY-ARG-PRO-ARG-THR-THR-ZXW-PHE-ALA-GLU, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | Authors: | Chu, N, Gabelli, S.B, Cole, P.A. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-22 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
7JUK
 
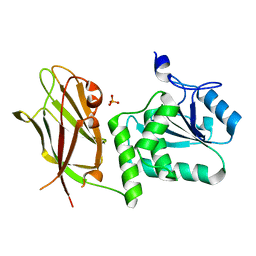 | | Crystal structure of PTEN with a tetra-phosphorylated tail (4p-crPTEN-13sp-T2, SDTTDSDPENEG) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6VJC
 
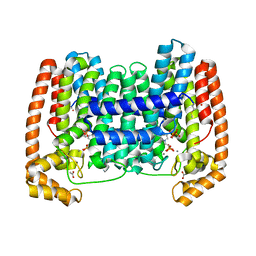 | | LmFPPS mutant T164Y in complex with 476A, IPP & Ca | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Aripirala, S, Gabelli, S.B. | | Deposit date: | 2020-01-15 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
6W7I
 
 | | LmFPPS mutant T164W in complex with 476A, IPP & Ca | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Aripirala, S, Gabelli, S.B. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
6WW1
 
 | | Crystal structure of the LmFPPS mutant E97Y | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Gabelli, S.B. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
6NCT
 
 | | Structure of p110alpha/niSH2 - vector data collection | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | Authors: | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
