2R9S
 
 | | c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 10, N-(tert-butyl)-4-[5-(pyridin-2-ylamino)quinolin-3-yl]benzenesulfonamide, ... | | Authors: | Habel, J. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1VKC
 
 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
3KVX
 
 | | JNK3 bound to aminopyrimidine inhibitor, SR-3562 | | Descriptor: | Mitogen-activated protein kinase 10, N-[(2Z)-4-(3-fluoro-5-morpholin-4-ylphenyl)pyrimidin-2(1H)-ylidene]-4-(3-morpholin-4-yl-1H-1,2,4-triazol-1-yl)aniline | | Authors: | Habel, J.E, Laughlin, J.D, LoGrasso, P. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, Biological Evaluation, X-ray Structure, and Pharmacokinetics of Aminopyrimidine c-jun-N-terminal Kinase (JNK) Inhibitors
J.Med.Chem., 53, 2010
|
|
1ZD0
 
 | | Crystal structure of Pfu-542154 conserved hypothetical protein | | Descriptor: | MAGNESIUM ION, METHANOL, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J.E, Liu, Z.J, Horanyi, P.S, Florence, Q.J.T, Tempel, W, Zhou, W, Chen, L, Lee, D, Nguyen, J, Chang, S.H, Bereton, P, Izumi, M, Jenny Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-13 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pfu-542154 conserved hypothetical protein
To be Published
|
|
5UVM
 
 | | HIT family hydrolase from Clostridium thermocellum Cth-393 | | Descriptor: | ADENOSINE, Histidine triad (HIT) protein, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J, Tempel, W, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIT family hydrolase from Clostridium thermocellum Cth-393
To Be Published
|
|
1YB3
 
 | | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus | | Descriptor: | UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Habel, J, Zhou, W, Chang, J, Zhao, M, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-19 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus
To be published
|
|
3FI3
 
 | | Crystal structure of JNK3 with indazole inhibitor, SR-3737 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(2-fluorophenyl)amino]-1H-indazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E, Duckett, D, LoGrasso, P. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
3FI2
 
 | | Crystal structure of JNK3 with amino-pyrazole inhibitor, SR-3451 | | Descriptor: | 1,2-ETHANEDIOL, 3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
3FV8
 
 | | JNK3 bound to piperazine amide inhibitor, SR2774. | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(3-chloro-2-(4-(prop-2-ynyl)piperazin-1-yl)phenyl)furan-2-carboxamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2009-01-15 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Synthesis and SAR of piperazine amides as novel c-jun N-terminal kinase (JNK) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1PVP
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, ALSHG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | Descriptor: | 34-MER, Recombinase cre | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVQ
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | Descriptor: | DNA 34-MER, Recombinase cre | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVR
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE LOXP (WILDTYPE) RECOGNITION SITE | | Descriptor: | 34-MER, Recombinase CRE | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
7T27
 
 | | Structure of phage FBB1 anti-CBASS nuclease Acb1-3'3'-cGAMP complex in post reaction state | | Descriptor: | Acb1, SULFATE ION, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphoryl]oxymethyl]-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphinic acid | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T26
 
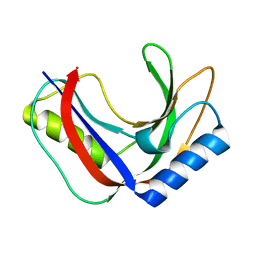 | | Structure of phage FBB1 anti-CBASS nuclease Acb1 in apo state | | Descriptor: | Acb1 | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T28
 
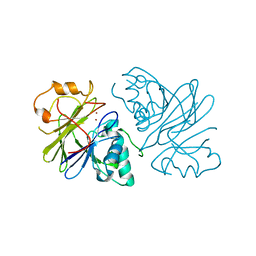 | | Structure of phage Bsp38 anti-Pycsar nuclease Apyc1 in apo state | | Descriptor: | Putative metal-dependent hydrolase, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7U2R
 
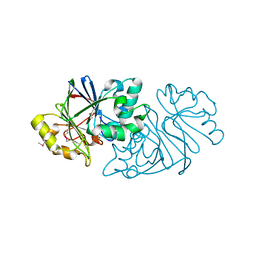 | | Structure of Paenibacillus sp. J14 Apyc1 | | Descriptor: | Apyc1, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-02-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7U2S
 
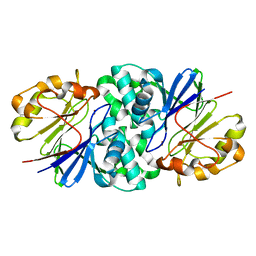 | | Structure of Paenibacillus xerothermodurans Apyc1 in the apo state | | Descriptor: | Apyc1, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-02-24 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
1DMK
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
1DMJ
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 5,6-CYCLIC-TETRAHYDROPTERIDINE | | Descriptor: | 7-AMINO-3,3A,4,5-TETRAHYDRO-8H-2-OXA-5,6,8,9B-TETRAAZA-CYCLOPENTA[A]NAPHTHALENE-1,9-DIONE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
1XMA
 
 | | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833 | | Descriptor: | MERCURY (II) ION, Predicted transcriptional regulator, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chen, L, Lee, D, Habel, J, Nguyen, J, Chang, S.-H, Kataeva, I, Xu, H, Chang, J, Zhao, M, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Praissman, J, Zhang, H, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833
To be published
|
|
1XRG
 
 | | Conserved hypothetical protein from Clostridium thermocellum Cth-2968 | | Descriptor: | Putative translation initiation inhibitor, yjgF family, UNKNOWN ATOM OR ION | | Authors: | Zhao, M, Chang, J, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Zhang, H, Arendall III, W.B, Ljundahl, L, Liu, Z.-J, Rose, J, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-14 | | Release date: | 2004-12-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved hypothetical protein from Clostridium thermocellum Cth-2968
To be published
|
|
1XG7
 
 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
1XHO
 
 | | Chorismate mutase from Clostridium thermocellum Cth-682 | | Descriptor: | Chorismate mutase, UNKNOWN ATOM OR ION | | Authors: | Xu, H, Chen, L, Lee, D, Habel, J.E, Nguyen, J, Chang, S.-H, Kataeva, I, Chang, J, Zhao, M, Yang, H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Away from the edge II: in-house Se-SAS phasing with chromium radiation.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y81
 
 | | Conserved hypothetical protein Pfu-723267-001 from Pyrococcus furiosus | | Descriptor: | COENZYME A, THIOCYANATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Zhao, M, Chang, J, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Conserved hypothetical protein Pfu-723267-001 from Pyrococcus furiosus
To be published
|
|
1XI3
 
 | | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Thiamine phosphate pyrophosphorylase, ... | | Authors: | Zhou, W, Chen, L, Tempel, W, Liu, Z.-J, Habel, J, Lee, D, Lin, D, Chang, S.-H, Dillard, B.D, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Kelley, L.-L.C, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiamine phosphate pyrophosphorylase from Pyrococcus furiosus Pfu-1255191-001
To be published
|
|
