2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | Descriptor: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
2YGL
 
 | | The X-ray crystal structure of tandem CBM51 modules of Sp3GH98, the family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
2YA0
 
 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2XYS
 
 | | Crystal structure of Aplysia californica AChBP in complex with strychnine | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, STRYCHNINE | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2XYT
 
 | | Crystal structure of Aplysia californica AChBP in complex with d- tubocurarine | | Descriptor: | D-TUBOCURARINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2WMG
 
 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in complex with the LewisY pentasaccharide blood group antigen. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2WMF
 
 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2X5V
 
 | | 80 microsecond laue diffraction snapshot from crystals of a photosynthetic reaction centre 3 millisecond following photoactivation. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-Induced Structural Changes in a Photosynthetic Reaction Center Caught by Laue Diffraction.
Science, 328, 2010
|
|
2WMK
 
 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the A-LewisY pentasaccharide blood group antigen. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2XQX
 
 | |
2YBA
 
 | | Crystal structure of Nurf55 in complex with histone H3 | | Descriptor: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
2YB8
 
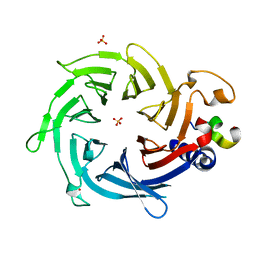 | | Crystal structure of Nurf55 in complex with Su(z)12 | | Descriptor: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
3A59
 
 | |
2ZP6
 
 | |
6HOL
 
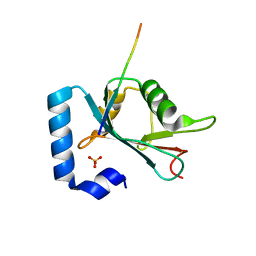 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | Descriptor: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOH
 
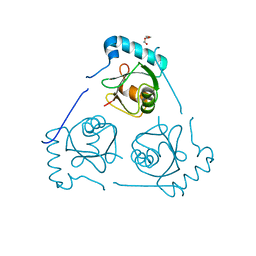 | | Structure of VPS34 LIR motif (S249E) bound to GABARAP | | Descriptor: | Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, TRIETHYLENE GLYCOL | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
7YZW
 
 | |
6OG7
 
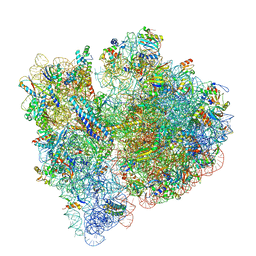 | | 70S termination complex with RF2 bound to the UGA codon. Non-rotated ribosome with RF2 bound (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OPM
 
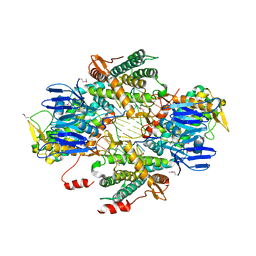 | | Casposase bound to integration product | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, DNA 21-mer, ... | | Authors: | Dyda, F, Hickman, A.B, Kailasan, S. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Elife, 9, 2020
|
|
6ORG
 
 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
6OGF
 
 | | 70S termination complex with RF2 bound to the UGA codon. Partially rotated ribosome with RF2 bound (Structure III). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OHY
 
 | | Chimpanzee SIV Env trimeric ectodomain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallesen, J, Andrabi, R, de Val, N, Burton, D.R, Ward, A.B. | | Deposit date: | 2019-04-08 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Chimpanzee SIV Envelope Trimer: Structure and Deployment as an HIV Vaccine Template.
Cell Rep, 27, 2019
|
|
6ORF
 
 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, SpGH29, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
6OGI
 
 | | 70S termination complex with RF2 bound to the UAG codon. Rotated ribosome conformation (Structure V) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6P62
 
 | | HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env BG505 NFL TD+, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
