9BUJ
 
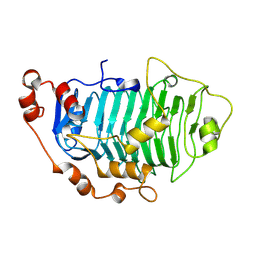 | | Structure of PfPL1 from Pseudoalteromonas fuliginea | | Descriptor: | CALCIUM ION, Pectate lyase | | Authors: | Boraston, A.B, Hobbs, J.K. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structure of a pectin-active family 1 polysaccharide lyase from the marine bacterium Pseudoalteromonas fuliginea.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9BDP
 
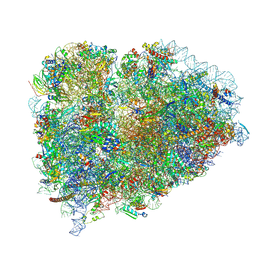 | |
9BDN
 
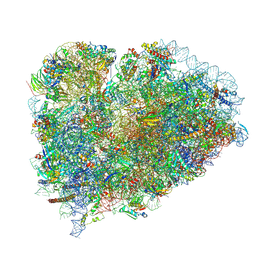 | |
9FJK
 
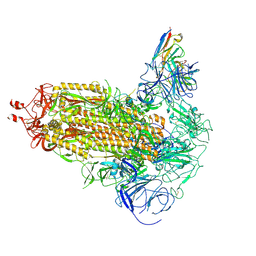 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | Descriptor: | K501SP6 Fv Heavy Chain, K501SP6 Fv Light Chain, Spike glycoprotein,Fibritin | | Authors: | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | Deposit date: | 2024-05-31 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6
To Be Published
|
|
2YA1
 
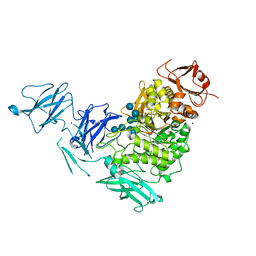 | | Product complex of a multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | PUTATIVE ALKALINE AMYLOPULLULANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
2YHG
 
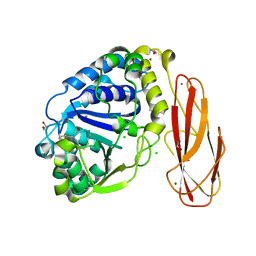 | | Ab initio phasing of a nucleoside hydrolase-related hypothetical protein from Saccharophagus degradans that is associated with carbohydrate metabolism | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Hehemann, J.H, Marsters, C, Boraston, A.B. | | Deposit date: | 2011-04-30 | | Release date: | 2011-08-03 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ab initio phasing of a nucleoside hydrolase-related hypothetical protein from Saccharophagus degradans that is associated with carbohydrate metabolism.
Proteins, 79, 2011
|
|
2YME
 
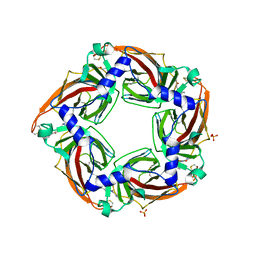 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
4MW2
 
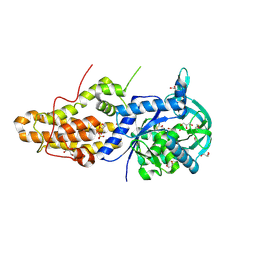 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[5-chloro-2-hydroxy-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1472) | | Descriptor: | 1-(3-{[5-chloro-2-hydroxy-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW7
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1469) | | Descriptor: | 1-{3-[(5-chloro-2-ethoxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW0
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea (Chem 1392) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MW6
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[2-(benzyloxy)-5-chloro-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1476) | | Descriptor: | 1-(3-{[2-(benzyloxy)-5-chloro-3-(prop-2-en-1-yl)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4N3U
 
 | | Candida albicans Superoxide Dismutase 5 (SOD5), Cu(II) | | Descriptor: | CITRATE ANION, COPPER (II) ION, Potential secreted Cu/Zn superoxide dismutase | | Authors: | Galaleldeen, A, Taylor, A.B, Waninger-Saroni, J.J, Holloway, S.P, Hart, P.J. | | Deposit date: | 2013-10-07 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2YGM
 
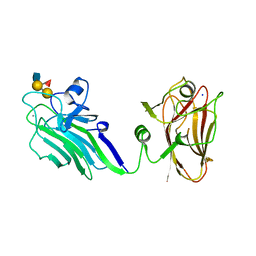 | | THE X-RAY CRYSTAL STRUCTURE OF TANDEM CBM51 MODULES OF SP3GH98, THE FAMILY 98 GLYCOSIDE HYDROLASE FROM STREPTOCOCCUS PNEUMONIAE SP3-BS71, IN COMPLEX WITH THE BLOOD GROUP B ANTIGEN | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION, SODIUM ION, ... | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
2YA2
 
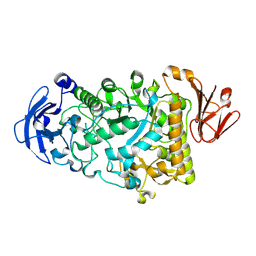 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA in complex with an inhibitor. | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2QWQ
 
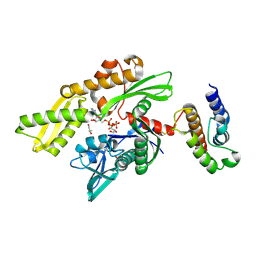 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-394aa)R171C and bovine Auxilin (810-910aa)D876C in the AMPPNP hydrolyzed form | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
4MW5
 
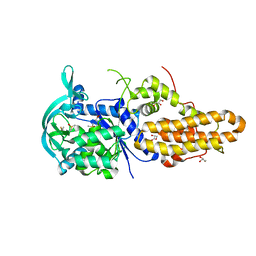 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-phenylurea (Chem 1415) | | Descriptor: | 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-phenylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MWD
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1433) and ATP analog AMPPCP | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
2WCS
 
 | | Crystal Structure of Debranching enzyme from Nostoc punctiforme (NPDE) | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Dumbrepatil, A.B, Choi, J.H, Nam, S.H, Park, K.H, Woo, E.J. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
2WC0
 
 | | crystal structure of human insulin degrading enzyme in complex with iodinated insulin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Manolopoulou, M, Guo, Q, Malito, E, Schilling, A.B, Tang, W.J. | | Deposit date: | 2009-03-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Basis of Catalytic Chamber-Assisted Unfolding and Cleavage of Human Insulin by Human Insulin Degrading Enzyme.
J.Biol.Chem., 284, 2009
|
|
2W8F
 
 | | Aplysia californica AChBP bound to in silico compound 31 | | Descriptor: | (3-EXO)-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8,8-DIMETHYL-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
2XM3
 
 | | Deinococcus radiodurans ISDra2 Transposase Left end DNA complex | | Descriptor: | 5'-D(*TP*TP*AP*GP*T)-3', ACETATE ION, DRA2 TRANSPOSASE BINDING ELEMENT, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XOM
 
 | | Atomic resolution structure of TmCBM61 in complex with beta-1,4- galactotriose | | Descriptor: | ARABINOGALACTAN ENDO-1,4-BETA-GALACTOSIDASE, CALCIUM ION, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Cid, M, Lodberg-Pedersen, H, Kaneko, S, Coutinho, P.M, Henrissat, B, Willats, W.G.T, Boraston, A.B. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Recognition of the Helical Structure of Beta-1,4-Galactan by a New Family of Carbohydrate-Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XZ5
 
 | | MMTS-modified Y53C mutant of Aplysia AChBP in complex with acetylcholine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE, ... | | Authors: | Brams, M, Gay, E.A, Colon Saez, J, Guskov, A, van Elk, R, van der Schors, R.C, Peigneur, S, Tytgat, J, Strelkov, S.V, Smit, A.B, Yakel, J.L, Ulens, C. | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of a Cysteine-Modified Mutant in Loop D of Acetylcholine Binding Protein
J.Biol.Chem., 286, 2011
|
|
2WMK
 
 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the A-LewisY pentasaccharide blood group antigen. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
