8DZ4
 
 | |
8DYY
 
 | |
7T73
 
 | | HIV-1 Envelope ApexGT2.2MUT in complex with PCT64.LMCA Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV Envelope ApexGT2.2MUT gp120, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Human immunoglobulin repertoire analysis guides design of vaccine priming immunogens targeting HIV V2-apex broadly neutralizing antibody precursors.
Immunity, 55, 2022
|
|
8DZ3
 
 | |
7T9A
 
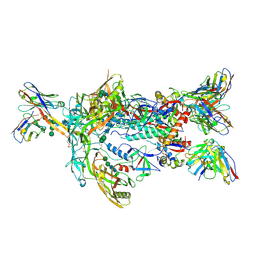 | | ApexGT2 in complex with GT2-d42.16 and RM20A3 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GT2-d42.16 Fab Heavy Chain, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2021-12-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Priming HIV Envelope V2 Apex-directed broadly neutralizing antibody responses with protein or mRNA immunogens
Immunity, 2022
|
|
8E6K
 
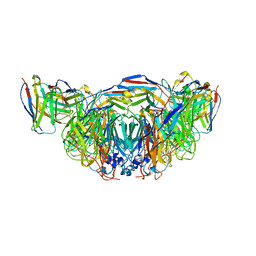 | | 2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2H08 fragment antigen binding heavy chain, 2H08 fragment antigen binding light chain, ... | | Authors: | Turner, H.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
2JE3
 
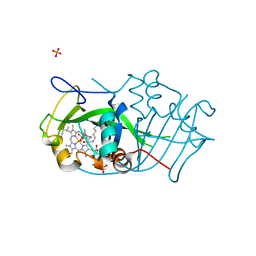 | | Cytochrome P460 from Nitrosomonas europaea - probable physiological form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
8E6J
 
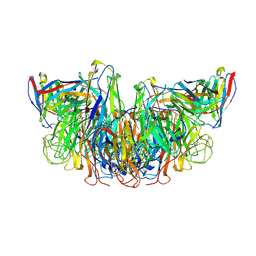 | |
2JE2
 
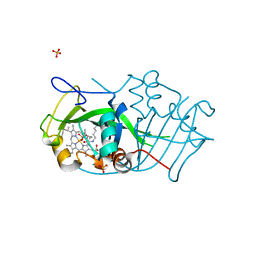 | | Cytochrome P460 from Nitrosomonas europaea - probable nonphysiological oxidized form | | Descriptor: | CYTOCHROME P460, HEME C, PHOSPHATE ION | | Authors: | Pearson, A.R, Elmore, B.O, Yang, C, Ferrara, J.D, Hooper, A.B, Wilmot, C.M. | | Deposit date: | 2007-01-13 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Cytochrome P460 of Nitrosomonas Europaea Reveals a Novel Cytochrome Fold and Heme-Protein Cross-Link.
Biochemistry, 46, 2007
|
|
5AC5
 
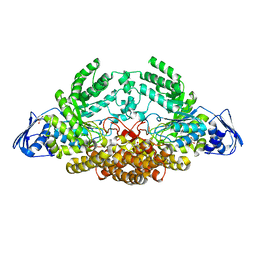 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
2JD9
 
 | |
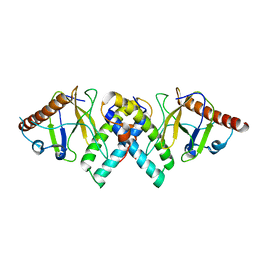
5AKO
 
 | |
5ABJ
 
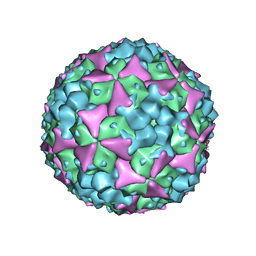 | | Structure of Coxsackievirus A16 in complex with GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
1HR7
 
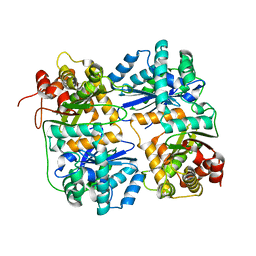 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
5ZBU
 
 | |
2KTU
 
 | |
5A5A
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with PNP-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
6B0C
 
 | | KLP10A-AMPPNP in complex with curved tubulin and a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
5A56
 
 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights into Its Mechanism.
J.Biol.Chem., 290, 2015
|
|
6B0L
 
 | | KLP10A-AMPPNP in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
5ACO
 
 | | Cryo-EM structure of PGT128 Fab in complex with BG505 SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 ENVELOPE GLYCOPROTEIN, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy.
Structure, 23, 2015
|
|
6APQ
 
 | | Anti-Marburgvirus Nucleoprotein Single Domain Antibody B | | Descriptor: | Anti-Marburgvirus Nucleoprotein Single Domain Antibody B, CHLORIDE ION, SODIUM ION | | Authors: | Taylor, A.B, Garza, J.A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling a Drift Resistant Cryptotope withinMarburgvirusNucleoprotein Recognized by Llama Single-Domain Antibodies.
Front Immunol, 8, 2017
|
|
2KTV
 
 | |
5A29
 
 | | Family 2 Pectate Lyase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, EXOPOLYGALACTURONATE LYASE, MANGANESE (II) ION, ... | | Authors: | McLean, R, Hobbs, J.K, Suits, M.D, Tuomivaara, S, Jones, D, Boraston, A.B, Abbott, D.W. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Analyses of Resurrected and Contemporary Enzymes Illuminate an Evolutionary Path for the Emergence of Exolysis in Polysaccharide Lyase Family 2.
J.Biol.Chem., 290, 2015
|
|
1UX7
 
 | | Carbohydrate-Binding Module CBM36 in complex with calcium and xylotriose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, SULFATE ION, ... | | Authors: | Davies, G.J, Boraston, A.B, Jamal, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
