6EID
 
 | | Crystal structure of wild-type Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, PHOSPHATE ION, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
3O2P
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3O4Q
 
 | |
5XDQ
 
 | | Bovine heart cytochrome c oxidase in the fully oxidized state with pH 7.3 at 1.77 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-03-29 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of bovine cytochrome c oxidase crystallized at a neutral pH using a fluorinated detergent.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5BTP
 
 | | Fusobacterium ulcerans ZTP riboswitch bound to ZMP | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Recognition of the bacterial alarmone ZMP through long-distance association of two RNA subdomains.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7TV9
 
 | | HUMAN COMPLEMENT COMPONENT C3B IN COMPLEX WITH APL-1030 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APL-1030 Nanofitin, Complement C3 beta chain, ... | | Authors: | Fontano, E, Nadupalli, A, Lakshminarasimhan, D, White, A, Garlish, J, Cinier, M, Chevrel, A, Perrocheau, A, Eyerman, D, Orme, M, Kitten, O, Scheibler, L. | | Deposit date: | 2022-02-04 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of APL-1030, a Novel, High-Affinity Nanofitin Inhibitor of C3-Mediated Complement Activation.
Biomolecules, 12, 2022
|
|
6BKR
 
 | |
3O4Y
 
 | | Crystal structure of CAD domain of the Plasmodium Vivax CDPK, PVX_11610 | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase 3, GLYCEROL, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of CAD domain of the Plasmodium Vivax CDPK, PVX_11610
TO BE PUBLISHED
|
|
3O5F
 
 | | Fk1 domain of FKBP51, crystal form VII | | Descriptor: | CARBONATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5M
 
 | | Fk1 domain mutant A19T of FKBP51, crystal form II | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5L
 
 | | Fk1 domain mutant A19T of FKBP51, crystal form I | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5Q
 
 | | Fk1 domain mutant A19T of FKBP51, crystal form IV, in presence of DMSO | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5GAT
 
 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
7TFP
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (1S,3S)-3-amino-4-(difluoromethylene)cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-amino-4-(fluoromethyl)cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-06 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
5L3E
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5A8X
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutrophil elastase, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | vonNussbaum, F, Li, V.M, Meibom, D, Anlauf, S, Bechem, M, Delbeck, M, Gerisch, M, Harrenga, A, Karthaus, D, Lang, D, Lustig, K, Mittendorf, J, Schaefer, M, Schaefer, S, Schamberger, J. | | Deposit date: | 2015-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Potent and Selective Human Neutrophil Elastase Inhibitors with Novel Equatorial Ring Topology: in vivo Efficacy of the Polar Pyrimidopyridazine BAY-8040 in a Pulmonary Arterial Hypertension Rat Model.
ChemMedChem, 11, 2016
|
|
3O83
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3OB4
 
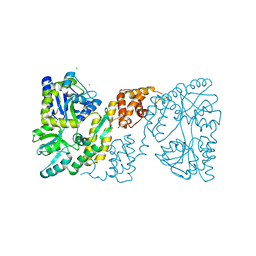 | | MBP-fusion protein of the major peanut allergen Ara h 2 | | Descriptor: | CHLORIDE ION, Maltose ABC transporter periplasmic protein,Arah 2, SULFATE ION, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Moon, A.F, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Allergy, 66, 2011
|
|
7TIK
 
 | |
5APO
 
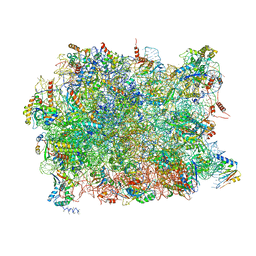 | | Structure of the yeast 60S ribosomal subunit in complex with Arx1, Alb1 and C-terminally tagged Rei1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Greber, B.J, Gerhardy, S, Leitner, A, Leibundgut, M, Salem, M, Boehringer, D, Leulliot, N, Aebersold, R, Panse, V.G, Ban, N. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation.
Cell(Cambridge,Mass.), 164, 2016
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
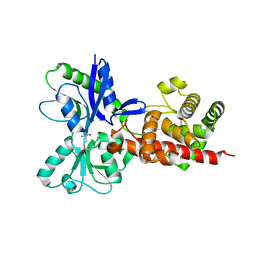
5AA3
 
 | |
6GGF
 
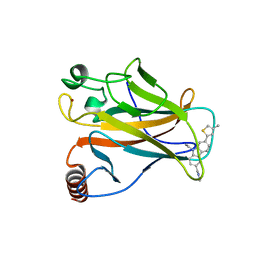 | |
6GHV
 
 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
6GJ7
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH 22 | | Descriptor: | (3~{S})-5-oxidanyl-3-[2-[[[1-(phenylmethyl)indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
