5F2D
 
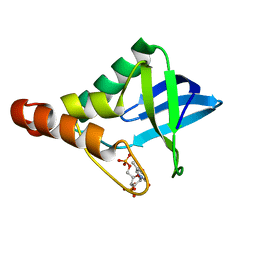 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92N/V99T at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92N/V99T at cryogenic temperature
To be Published
|
|
7OW7
 
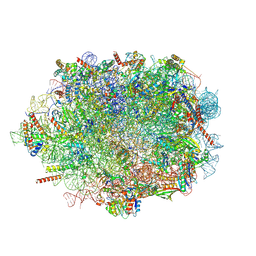 | |
5I19
 
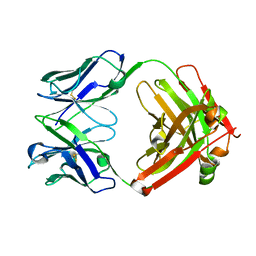 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV3-23/IGKV1-39 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
7A9A
 
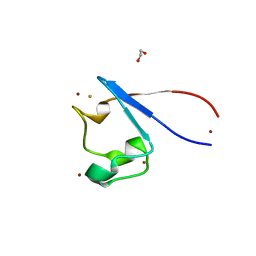 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
5I1J
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV5-51/IGKV3-11 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
1D62
 
 | |
4HZG
 
 | | Structure of haloalkane dehalogenase DhaA from Rhodococcus rhodochrous | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Stsiapanava, A, Weiss, M.S, Mesters, J.R, Kuta Smatanova, I. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4XUQ
 
 | |
6ORL
 
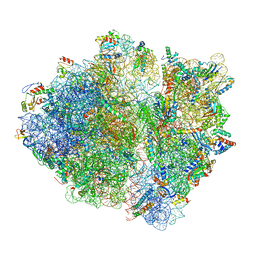 | | RF1 pre-accommodated 70S complex at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ZPL
 
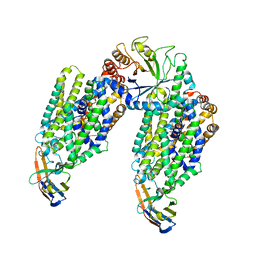 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
6XLS
 
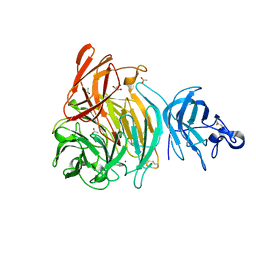 | |
5IAI
 
 | | Crystal structure of ABC transporter Solute Binding Protein Arad_9887 from Agrobacterium radiobacter K84, target EFI-510945 in complex with Ribitol | | Descriptor: | D-ribitol, GLYCEROL, Sugar ABC transporter | | Authors: | Vetting, M.W, Bonanno, J.B, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein Arad_9887 from Agrobacterium radiobacter K84, target EFI-510945 in complex with Ribitol
To be published
|
|
3EAE
 
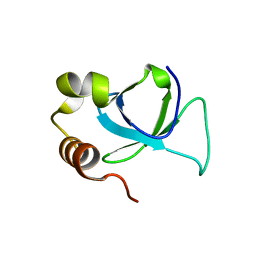 | | PWWP domain of human hepatoma-derived growth factor 2 (HDGF2) | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Amaya, M.F, Zeng, H, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
5LLP
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 3-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
2DEG
 
 | | Crystal structure of tt0972 protein form Thermus Thermophilus with Mn2(+) ions | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Inagaki, E, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-10 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tt0972 protein form Thermus Thermophilus
To be Published
|
|
6XPM
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature | | Descriptor: | Lysophospholipase L1, SODIUM ION | | Authors: | Kim, Y, Johnson, J, Welk, L, Endres, M, Levens, A, Sherrell, D.A, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature
To Be Published
|
|
8TWD
 
 | | Structure of IraM-bound RssB | | Descriptor: | ACETATE ION, Anti-adapter protein IraM, Regulator of RpoS | | Authors: | Brugger, C, Deaconescu, A.M. | | Deposit date: | 2023-08-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | IraM remodels the RssB segmented helical linker to stabilize sigma s against degradation by ClpXP.
J.Biol.Chem., 300, 2023
|
|
6HI2
 
 | | PI3 Kinase Delta in complex with 3{6[(1S,6R)3oxabicyclo[4.1.0]heptan6yl]pyridin2yl}phenol | | Descriptor: | 3-[6-[(1~{S},6~{R})-3-oxabicyclo[4.1.0]heptan-6-yl]pyridin-2-yl]phenol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Summers, D, Peace, S. | | Deposit date: | 2018-08-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A theoretical and experimental investigation into the conformational bias of aryl cyclopropylpyrans, novel bioisosteres for N-aryl morpholines.
to be published
|
|
6ROB
 
 | | Human Carbonic Anhydrase II in complex with 4-cyanobenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-cyanobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.929 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
8AJQ
 
 | |
5LUN
 
 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - P1 ultra-high resolution crystal form in complex with iron, N-oxalylglycine and arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, FE (III) ION, ... | | Authors: | McDonough, M.A, Zhang, Z, Schofield, C.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5I6F
 
 | | Crystal structure of C-terminal variant 1 of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
5F7E
 
 | | Crystal structure of germ-line precursor of 3BNC60 Fab | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Sievers, S.A, Scharf, L, Jiang, S, Bjorkman, P.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
6U2T
 
 | |
