6L5Y
 
 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 140 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TPK
 
 | | Crystal structure of the human oxytocin receptor | | Descriptor: | (3~{R},6~{R})-6-[(2~{S})-butan-2-yl]-3-(2,3-dihydro-1~{H}-inden-2-yl)-1-[(1~{R})-1-(2-methyl-1,3-oxazol-4-yl)-2-morpholin-4-yl-2-oxidanylidene-ethyl]piperazine-2,5-dione, CHOLESTEROL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waltenspuehl, Y, Schoeppe, J, Ehrenmann, J, Kummer, L, Plueckthun, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human oxytocin receptor.
Sci Adv, 6, 2020
|
|
6XHL
 
 | | Covalent complex of SARS-CoV main protease with N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Gajiwala, K.S, Ferre, R.A, Ryan, K, Stewart, A.E. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19.
J.Med.Chem., 63, 2020
|
|
5F3M
 
 | | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution . | | Descriptor: | 1,2-ETHANEDIOL, 7,8-dihydroneopterin aldolase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution .
To Be Published
|
|
8A1N
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with fumaryl amide analogue 13 | | Descriptor: | (Z)-N-(4-chlorophenyl)-4-oxidanylidene-but-2-enamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
4HJJ
 
 | | Structure Reveals Function of the Dual Variable Domain Immunoglobulin (DVD-Ig) Molecule | | Descriptor: | Anti-IL12 Anti-IL18 DFab Heavy Chain, Anti-IL12 Anti-IL18 DFab Light Chain, CHLORIDE ION, ... | | Authors: | Jakob, C.G, Edalji, R, Judge, R.A, DiGiammarino, E, Li, Y, Gu, J, Ghayur, T. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure reveals function of the dual variable domain immunoglobulin (DVD-Ig[TM]) molecule
MAbs, 5, 2013
|
|
6X9Y
 
 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
6KSS
 
 | |
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
8FWG
 
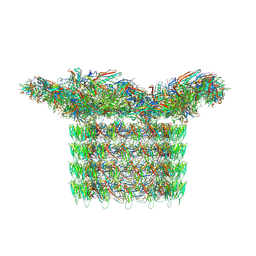 | | Structure of neck and portal vertex of Agrobacterium phage Milano, C5 symmetry | | Descriptor: | Collar sheath protein, gp13, Linking protein 1, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-22 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
8FWB
 
 | | Portal assembly of Agrobacterium phage Milano | | Descriptor: | Portal protein, gp7 | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
5XC6
 
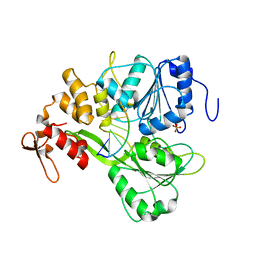 | | Dengue Virus 4 NS3 Helicase in complex with SSRNA SLA12 | | Descriptor: | NS3 Helicase, PHOSPHATE ION, RNA (5'-R(*AP*GP*UP*UP*GP*UP*UP*AP*GP*UP*CP*U)-3') | | Authors: | Swarbrick, C.M.D, Basavannacharya, C, Chan, K.W.K, Chan, S.A, Singh, D, Wei, N, Phoo, W.W, Luo, D, Lescar, J, Vasudevan, S.G. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication
Nucleic Acids Res., 45, 2017
|
|
1Z7U
 
 | |
6TUF
 
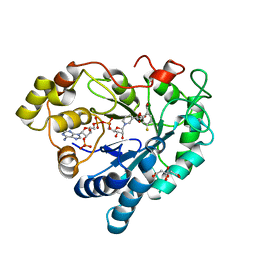 | | Human Aldose Reductase in complex with ALR43 | | Descriptor: | 2-[5-fluoranyl-2-[[3-[methyl(oxidanyl)-$l^{3}-sulfanyl]phenyl]methylcarbamoyl]phenoxy]ethanoic acid, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Human Aldose Reductase in complex with SAR25
To Be Published
|
|
8A29
 
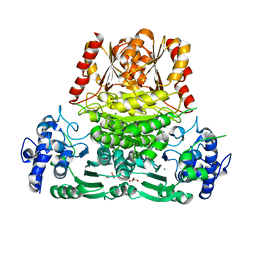 | | Apo 1-deoxy-D-xylulose 5-phosphate synthase from Pseudomonas aeruginosa | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hamid, R, Adam, S, Lacour, A, Monjas, L, Hirsch, A. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1-deoxy-D-xylulose-5-phosphate synthase from Pseudomonas aeruginosa and Klebsiella pneumoniae reveals conformational changes upon cofactor binding.
J.Biol.Chem., 299, 2023
|
|
8FWM
 
 | | Structure of tail-neck junction of Agrobacterium phage Milano | | Descriptor: | Collar sheath protein, gp13, Tail sheath protein, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
4ZU6
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus influenzae in complex with pre-reactive o-acetyl serine, alpha-aminoacrylate reaction intermediate and Peptide inhibitor at the resolution of 2.25A | | Descriptor: | C-terminal peptide from Serine acetyltransferase, Cysteine synthase, O-ACETYLSERINE, ... | | Authors: | Ekka, M.K, Singh, A.K, Kaushik, A, Kumaran, S. | | Deposit date: | 2015-05-15 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of O-Acetylserine Sulfhydrylase from Haemophilus inuen-zae in complex with pre-reactive o-acetyl serine and peptide inhibitor
To Be Published
|
|
8FWC
 
 | | Collar sheath structure of Agrobacterium phage Milano | | Descriptor: | Collar sheath protein, gp13 | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
4X9R
 
 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 3B | | Descriptor: | Phosphopeptide macrocycle 3B, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
1Z1F
 
 | | Crystal structure of stilbene synthase from Arachis hypogaea (resveratrol-bound form) | | Descriptor: | CITRIC ACID, RESVERATROL, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
6R3K
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[5-chloranyl-2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
7LVG
 
 | |
6ZKY
 
 | | Crystal structure of InhA:01 TCR in complex with HLA-E (S147C) bound to InhA (53-61 H3C) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
5IEX
 
 | | Crystal structure of (R,S)-S-{4-[(5-Bromo-4-{[(2R,3R)-2-hydroxy-1-methylpropyl]oxy}- pyrimidin-2-yl)amino]phenyl}-S-cyclopropylsulfoximide bound to CDK2 | | Descriptor: | (2R,3R)-3-[(5-bromo-2-{[4-(S-cyclopropylsulfonimidoyl)phenyl]amino}pyrimidin-4-yl)oxy]butan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
