8BPN
 
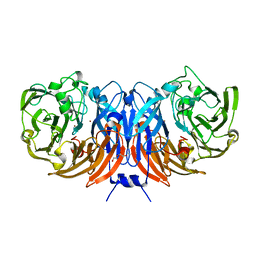 | | The structure of thiocyanate dehydrogenase mutant form with Phe 436 replaced by Gln from Thioalkalivibrio paradoxus | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Twin-arginine translocation signal domain-containing protein | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Kulikova, O.G, Dergousova, N.I, Rakitina, T.V, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-01-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Probing the Role of a Conserved Phenylalanine in the Active Site of Thiocyanate Dehydrogenase
Crystals, 12, 2022
|
|
1ASQ
 
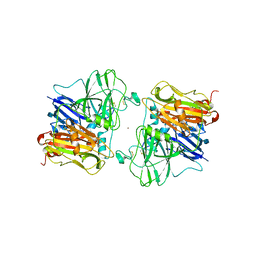 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
3LEZ
 
 | |
5M85
 
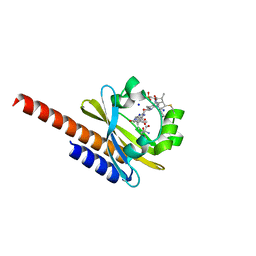 | | Three-dimensional structure of the intermediate state of GAF3 from Slr1393 of Synechocystis sp. PCC6803 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHYCOCYANOBILIN, ... | | Authors: | Xu, X.-L, Zhao, K.-H, Gaertner, W, Hoeppner, A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural elements regulating the photochromicity in a cyanobacteriochrome
Proc.Natl.Acad.Sci.USA, 2020
|
|
8RB4
 
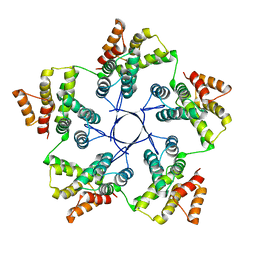 | |
3LOX
 
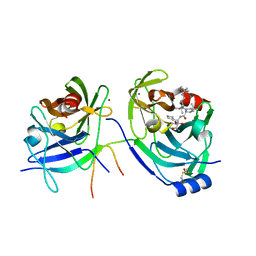 | | HCV NS3-4a protease domain with a ketoamide inhibitor derivative of Boceprevir bound | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-amino-1-cyclobutyl-3-hydroxy-4-oxobutan-2-yl]-6,6-dimethyl-3-{3-methyl-N-[(1-methylcyclohexyl)c arbamoyl]-L-valyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | Authors: | Prongay, A.J. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The introduction of P4 substituted 1-methylcyclohexyl groups into Boceprevir: a change in direction in the search for a second generation HCV NS3 protease inhibitor.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LG5
 
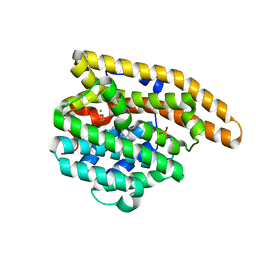 | | F198A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Aaron, J.A, Lin, X, Cane, D.E, Christianson, D.W. | | Deposit date: | 2010-01-19 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structure of Epi-Isozizaene Synthase from Streptomyces coelicolor A3(2), a Platform for New Terpenoid Cyclization Templates
Biochemistry, 49, 2010
|
|
6VBT
 
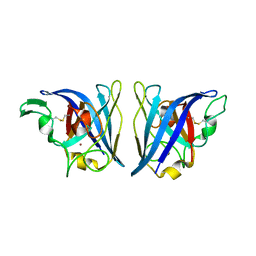 | |
5G2T
 
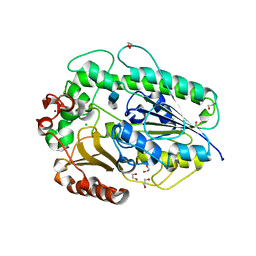 | | BT1596 in complex with its substrate 4,5 unsaturated uronic acid alpha 1,4 D-Glucosamine-2-N, 6-O-disulfate | | Descriptor: | 1,2-ETHANEDIOL, 2-O GLYCOSAMINOGLYCAN SULFATASE, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4QXX
 
 | |
5K9J
 
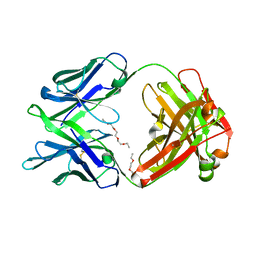 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 isolated following H5 immunization. | | Descriptor: | 56.a.09 heavy chain, 56.a.09 light chain, POLYETHYLENE GLYCOL (N=34) | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-31 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
6AN1
 
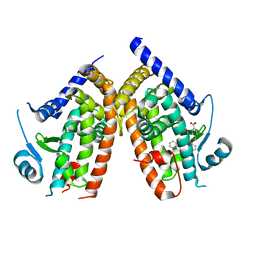 | | Crystal structure of the complex between PPARgamma LBD and the ligand AM-879 | | Descriptor: | 4-({2-[(1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)methyl]phenoxy}methyl)benzoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Veras, H, Figueira, A.C, le Maire, A. | | Deposit date: | 2017-08-11 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Screening for PPAR Non-Agonist Ligands Followed by Characterization of a Hit, AM-879, with Additional No-Adipogenic and cdk5-Mediated Phosphorylation Inhibition Properties.
Front Endocrinol (Lausanne), 9, 2018
|
|
8JSH
 
 | |
5G5N
 
 | | Structure of the snake adenovirus 1 hexon-interlacing LH3 protein, methylmercury chloride derivative | | Descriptor: | CHLORIDE ION, GLYCEROL, LH3 HEXON-INTERLACING CAPSID PROTEIN, ... | | Authors: | Nguyen, T.H, Singh, A.K, Albala-Perez, B, van Raaij, M.J. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid.
Structure, 25, 2017
|
|
5MWG
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ091 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-2,3-bis(oxidanylidene)-7-piperidin-1-yl-quinoxalin-6-yl]-5,6,7,8-tetrahydronaphthalene-2-sulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
6VDO
 
 | |
7MR8
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-076 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]benzoyl}-L-glutamic acid | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-076
To Be Published
|
|
8S9W
 
 | | Murine S100A7/S100A15 in presence of calcium | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Harrison, S.A, Naretto, A, Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-03-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Comparative analysis of the physical properties of murine and human S100A7: Insight into why zinc piracy is mediated by human but not murine S100A7.
J.Biol.Chem., 299, 2023
|
|
3LJF
 
 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
6YI0
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.65 A in P41212 space group | | Descriptor: | Histidine triad nucleotide-binding protein 2, mitochondrial, SODIUM ION | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
5MCL
 
 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 18.4 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
7MQU
 
 | | The haddock model of GDP KRas in complex with promethazine using NMR chemical shift perturbations | | Descriptor: | (2R)-N,N-dimethyl-1-(10H-phenothiazin-10-yl)propan-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.P. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
4RBS
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, ACETIC ACID, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem
To be Published
|
|
8RE3
 
 | | Crystal Structure determination of Dye-decolorizing Peroxidase (DyP) mutant M190G from Deinoccoccus radiodurans | | Descriptor: | CALCIUM ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Salgueiro, B.A, Frade, K, Frazao, C, Matias, P, Moe, E. | | Deposit date: | 2023-12-10 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical, Biophysical, and Structural Analysis of an Unusual DyP from the Extremophile Deinococcus radiodurans.
Molecules, 29, 2024
|
|
6YNM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_096 | | Descriptor: | 1-[(2~{R},5~{S})-2-(3-chlorophenyl)-5-methyl-morpholin-4-yl]-3-methoxy-propan-2-ol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
