4DKD
 
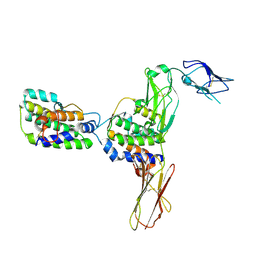 | | Crystal Structure of Human Interleukin-34 Bound to Human CSF-1R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-34, ... | | Authors: | Ma, X, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
4LV6
 
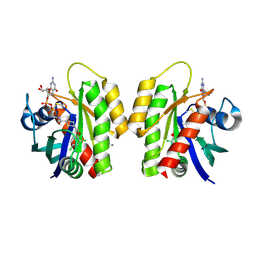 | | Crystal Structure of small molecule disulfide 4 covalently bound to K-Ras G12C | | Descriptor: | 1-[(2,4-dichlorophenoxy)acetyl]-N-(2-sulfanylethyl)piperidine-4-carboxamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-26 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
2P3C
 
 | | Crystal Structure of the subtype F wild type HIV protease complexed with TL-3 inhibitor | | Descriptor: | ACETIC ACID, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
2P43
 
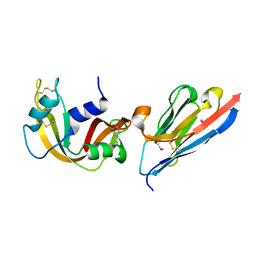 | | Complex of a camelid single-domain vhh antibody fragment with RNASE A at 1.65A resolution: SE3-mono-1 crystal form with three se-met sites (M34, M51, M83) in vhh scaffold | | Descriptor: | ANTIBODY CAB-RN05, Ribonuclease pancreatic | | Authors: | Tereshko, V, Uysal, S, Koide, A, Margalef, K, Koide, S, Kossiakoff, A.A. | | Deposit date: | 2007-03-11 | | Release date: | 2008-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold
Protein Sci., 17, 2008
|
|
3B3T
 
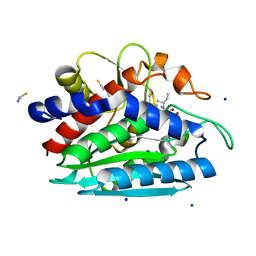 | | Crystal structure of the D118N mutant of the aminopeptidase from Vibrio proteolyticus | | Descriptor: | Bacterial leucyl aminopeptidase, ISOLEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
5MFW
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
5MDL
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its O2-derivatized form by a "soak-and-freeze" derivatization method | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4LZ0
 
 | | A236G Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LZC
 
 | | W325F Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
3B8M
 
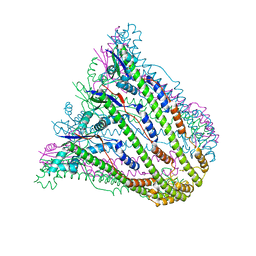 | |
1TPC
 
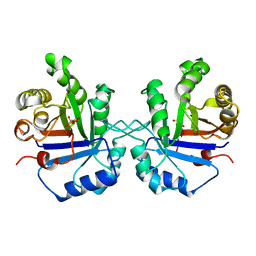 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
5MKU
 
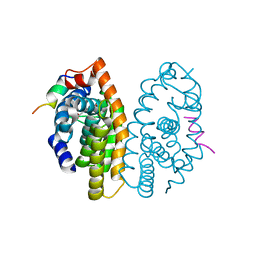 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 4 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(3-propan-2-ylphenyl)phenyl]prop-2-enoic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
1TPB
 
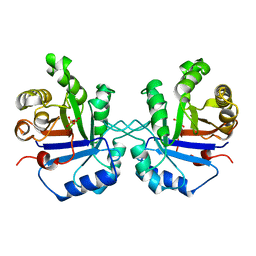 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
4DKC
 
 | | Crystal Structure of Human Interleukin-34 | | Descriptor: | Interleukin-34, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ma, X, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
5MNB
 
 | | Cationic trypsin in complex with 2-aminopyridine (deuterated sample at 295 K) | | Descriptor: | 2-AMINOPYRIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.939 Å) | | Cite: | Charges Shift Protonation: Neutron Diffraction Reveals that Aniline and 2-Aminopyridine Become Protonated Upon Binding to Trypsin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MNI
 
 | | Escherichia coli AGPase mutant R130A apo form | | Descriptor: | Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, Marina, A, Orrantia, A, Eguskiza, A, Guerin, M.E. | | Deposit date: | 2016-12-13 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanistic insights into the allosteric regulation of bacterial ADP-glucose pyrophosphorylases.
J. Biol. Chem., 292, 2017
|
|
4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
5XWT
 
 | | Crystal structure of PTPdelta Ig1-Fn1 in complex with SALM5 LRR-Ig | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.178 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
3BHY
 
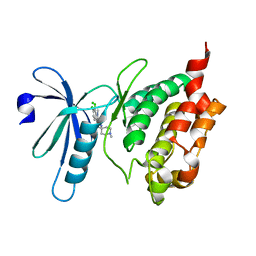 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with a beta-carboline ligand | | Descriptor: | (4R)-7,8-dichloro-1',9-dimethyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, CHLORIDE ION, Death-associated protein kinase 3 | | Authors: | Filippakopoulos, P, Rellos, P, Eswaran, J, Fedorov, O, Berridge, G, Niesen, F, Bracher, F, Huber, K, Pike, A.C.W, Roos, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | 7,8-dichloro-1-oxo-beta-carbolines as a versatile scaffold for the development of potent and selective kinase inhibitors with unusual binding modes.
J.Med.Chem., 55, 2012
|
|
4DSK
 
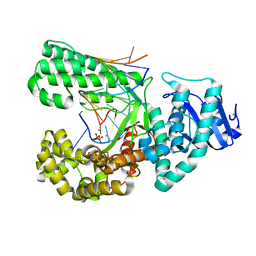 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, PPi and Calcium | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*CP*TP*AP*CP*AP*GP*GP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*AP*GP*TP*CP*CP*TP*GP*TP*AP*GP*CP*C)-3'), ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
3S3D
 
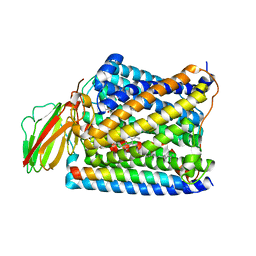 | | Structure of Thermus thermophilus cytochrome ba3 oxidase 480s after Xe depressurization | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Fee, J.A, Deniz, A.A, Stout, C.D. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba(3) oxidase.
Biochemistry, 51, 2012
|
|
2PFP
 
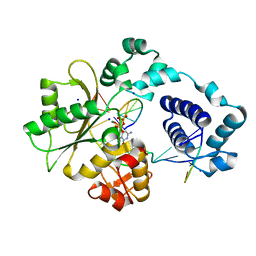 | | DNA Polymerase lambda in complex with DNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, Downstream Primer, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
3S5Z
 
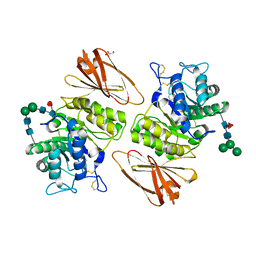 | | Pharmacological Chaperoning in Human alpha-Galactosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, GLYCEROL, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2011-05-23 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The molecular basis of pharmacological chaperoning in human alpha-galactosidase
Chem.Biol., 18, 2011
|
|
3S3U
 
 | |
4DSJ
 
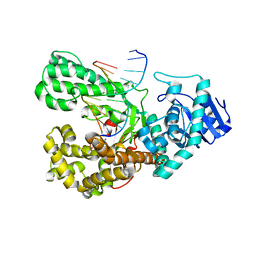 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, dGTP and Calcium | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA, ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
