4YTR
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
1DEO
 
 | | RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.55 A RESOLUTION WITH SO4 IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RHAMNOGALACTURONAN ACETYLESTERASE, SULFATE ION, ... | | Authors: | Molgaard, A, Kauppinen, S, Larsen, S. | | Deposit date: | 1999-11-15 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases.
Structure Fold.Des., 8, 2000
|
|
6I2N
 
 | | Helical RNA-bound Hantaan virus nucleocapsid | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*U)-3') | | Authors: | Arragain, B, Reguera, J, Desfosses, A, Gutsche, I, Schoehn, G, Malet, H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms.
Elife, 8, 2019
|
|
8QEV
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ornithine transcarbamylase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
6LNT
 
 | | Cryo-EM structure of immature Zika virus in complex with human antibody DV62.5 Fab | | Descriptor: | Envelope protein, Fab DV62.5 heavy-chain variable region, Fab DV62.5 light-chain variable region, ... | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
4Y3I
 
 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, low dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
6SBR
 
 | | The crystal structure of PfA-M1 in complex with 7-amino-1,4-dibromo-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Salomon, E, Schmitt, M, Mouray, E, McEwen, A.G, Torchy, M, Poussin-Courmontagne, P, Alavi, S, Tarnus, C, Cavarelli, J, Florent, I, Albrecht, S. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Aminobenzosuberone derivatives as PfA-M1 inhibitors: Molecular recognition and antiplasmodial evaluation.
Bioorg.Chem., 98, 2020
|
|
8E77
 
 | | rystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA), incomplete with its external aldimine reaction intermediate | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
6V4P
 
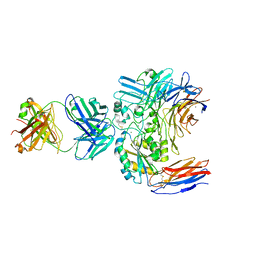 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | Descriptor: | Abciximab, heavy chain, light chain, ... | | Authors: | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
1DDL
 
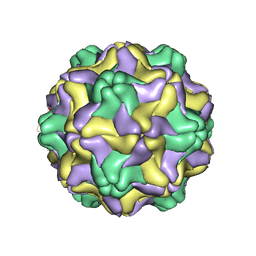 | | DESMODIUM YELLOW MOTTLE TYMOVIRUS | | Descriptor: | DESMODIUM YELLOW MOTTLE VIRUS, RNA (5'-R(P*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Larson, S.B, Day, J, Canady, M.A, Greenwood, A, McPherson, A. | | Deposit date: | 1999-11-10 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Refined structure of desmodium yellow mottle tymovirus at 2.7 A resolution.
J.Mol.Biol., 301, 2000
|
|
6I51
 
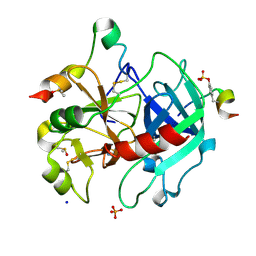 | | Thrombin in complex with fragment J02 | | Descriptor: | 1H-isoindol-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Abazi, N, Heine, A, Klebe, G. | | Deposit date: | 2018-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment screening of Thrombin using a 96 member fragment library
To Be Published
|
|
5JCI
 
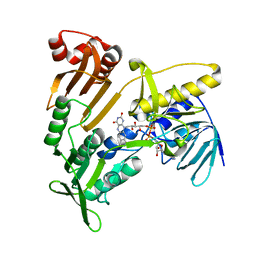 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Os09g0567300 protein | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
7R2G
 
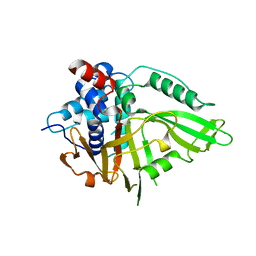 | | USP15 D1D2 in catalytically-competent state bound to mitoxantrone stack (isoform 2) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 15, ... | | Authors: | Priyanka, A, Sixma, T.K. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mitoxantrone stacking does not define the active or inactive state of USP15 catalytic domain.
J.Struct.Biol., 214, 2022
|
|
8AIH
 
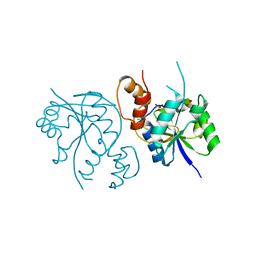 | | Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase at 1.9 Angstroms Resolution | | Descriptor: | DIHYDROGENPHOSPHATE ION, Probable nicotinate-nucleotide adenylyltransferase, SULFATE ION | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
5JCM
 
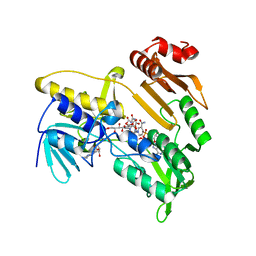 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ISOASCORBIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
8DO8
 
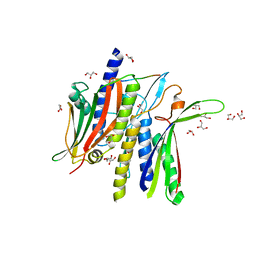 | | Crystal structure ATG9 HDIR in complex with the ATG13:ATG101 HORMA dimer | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13, GLYCEROL | | Authors: | Buffalo, C.Z, Ren, X, Yokom, A.L, Hurley, J.H. | | Deposit date: | 2022-07-12 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for ATG9A recruitment to the ULK1 complex in mitophagy initiation.
Sci Adv, 9, 2023
|
|
8E75
 
 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | Descriptor: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
1DL2
 
 | | CRYSTAL STRUCTURE OF CLASS I ALPHA-1,2-MANNOSIDASE FROM SACCHAROMYCES CEREVISIAE AT 1.54 ANGSTROM RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CLASS I ALPHA-1,2-MANNOSIDASE, ... | | Authors: | Vallee, F, Lipari, F, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 1999-12-08 | | Release date: | 2000-02-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of a class I alpha1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control.
EMBO J., 19, 2000
|
|
4YWD
 
 | | Structure of rat cytosolic pepck in complex with 2,3-Pyridine dicarboxylic acid | | Descriptor: | MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, cytosolic [GTP], ... | | Authors: | Balan, M.D, Johnson, T.A, Mcleod, M.J, Lotosky, W.R, Holyoak, T. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition and Allosteric Regulation of Monomeric Phosphoenolpyruvate Carboxykinase by 3-Mercaptopicolinic Acid.
Biochemistry, 54, 2015
|
|
8QEU
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with ornithine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-ornithine, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
8AGN
 
 | | Cyclohexane epoxide low pH soak of epoxide hydrolase from metagenomic source ch65 | | Descriptor: | (1R,6S)-7-oxabicyclo[4.1.0]heptane, 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
1DCI
 
 | | DIENOYL-COA ISOMERASE | | Descriptor: | 1,2-ETHANEDIOL, DIENOYL-COA ISOMERASE, MAGNESIUM ION, ... | | Authors: | Modis, Y, Filppula, S.A, Novikov, D, Norledge, B, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 1998-02-13 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis.
Structure, 6, 1998
|
|
8G33
 
 | |
6H05
 
 | | Cryo-electron microscopic structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate (2-oxoglutarate) dehydrogenase complex [residues 218-453] | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Nagy, B, Zambo, Z, Hubert, A, Polak, M, Nemeria, N.S, Novacek, J, Jordan, F, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-07-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: Implications for the overall hKGDHc structure.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
4Y5F
 
 | | PAS-GAF fragment from Deinococcus radiodurans BphP assembled with BV - Y307S, high dose | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, F, Burgie, E.S, Yu, T, Heroux, A, Schatz, G.C, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
J.Am.Chem.Soc., 137, 2015
|
|
