7S4I
 
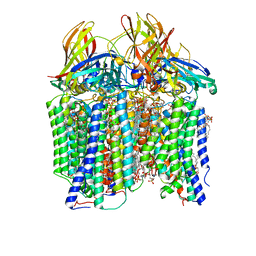 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.26 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
8ONN
 
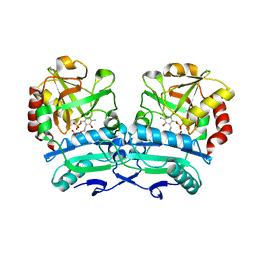 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | Descriptor: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
5LG3
 
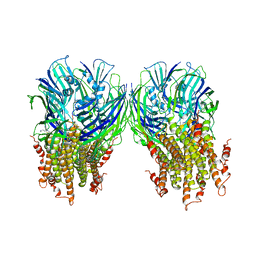 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.567 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6UBC
 
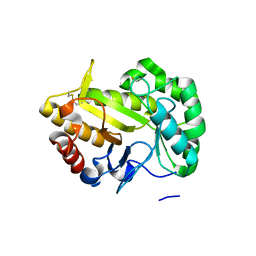 | | Crystal structure of a GH128 (subgroup VII) oligosaccharide-binding protein from Cryptococcus neoformans (CnGH128_VII) | | Descriptor: | Glyco_hydro_cc domain-containing protein | | Authors: | Santos, C.R, Costa, P.A.C.R, Souza, B.P, Vieira, P.S, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6ZNT
 
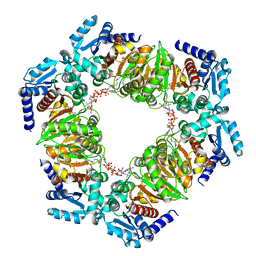 | | MaeB PTA domain, acetyl-CoA bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
8RTE
 
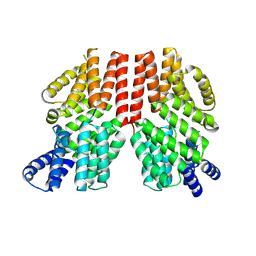 | |
7N67
 
 | | Crystal structure of HCAN_0198, a 3,4-ketoisomerase from Helicobacter canadensis | | Descriptor: | FdtA domain-containing protein, TETRAMETHYLAMMONIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Heisdorf, C.J, Griffiths, W.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of an unusual formylated sugar in the emerging human pathogen Helicobacter canadensis.
Protein Sci., 30, 2021
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6OT8
 
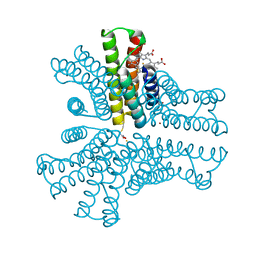 | | Bimetallic hexameric cage design 4 (BMC4) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
8RSU
 
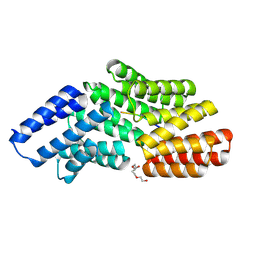 | |
8EQJ
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like membrane environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
6UEO
 
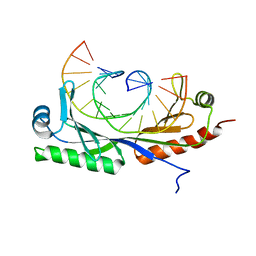 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
7NM8
 
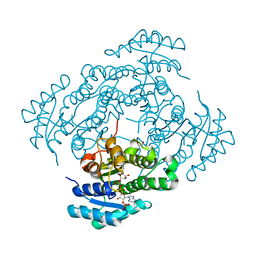 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM, in complex with NADPH | | Descriptor: | Antimycin pathway standalone ketoreductase, AntM, GLYCEROL, ... | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
6S5X
 
 | | Structure of RibR, the most N-terminal Rib domain from Group B Streptococcus species Streptococcus agalactiae | | Descriptor: | Group B streptococcal R4 surface protein, SODIUM ION | | Authors: | Whelan, F, Turkenburg, J.P, Griffiths, S.C, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LI7
 
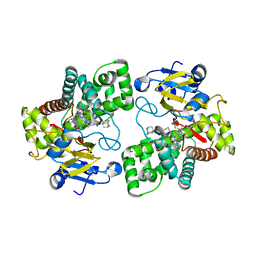 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with 1-(3-(1H-imidazol-1-yl)propyl)-3-((3s,5s,7s)-adamantan-1-yl)urea | | Descriptor: | 1-(1-adamantyl)-3-(3-imidazol-1-ylpropyl)urea, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
6TWH
 
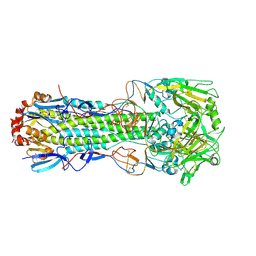 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Gly228Ser) from an H10N7 seal influenza virus isolated in Germany | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-13 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
8EQT
 
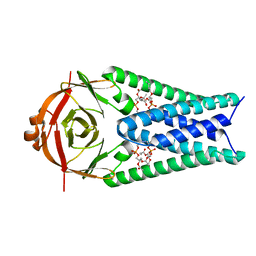 | | Structure of SARS-CoV-2 Orf3a in plasma membrane-like environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
7PL1
 
 | |
7NM7
 
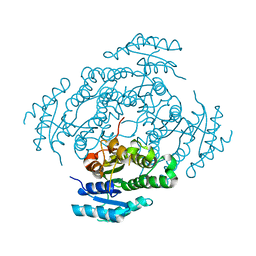 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM | | Descriptor: | Antimycin pathway standalone ketoreductase enzyme, AntM | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
8SDH
 
 | |
7PN0
 
 | | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Timofeev, V.I, Abramchik, Y.A, Kostromina, M.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2021-09-04 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group
To Be Published
|
|
8P5T
 
 | | Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
7SKK
 
 | | pertussis toxin in complex with ADPR and Nicotinamide | | Descriptor: | NICOTINAMIDE, Pertussis toxin subunit 1, SULFATE ION, ... | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2021-10-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65000772 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
6U64
 
 | | Mcl-1 bound to compound 17 | | Descriptor: | 5-[(2-phenylethyl)sulfanyl]-2-{[(4-phenylpiperazin-1-yl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6TWS
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Gly228Ser) from an H10N7 seal influenza virus isolated in Germany with 2mM EDTA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-13 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
