7NT7
 
 | |
7O2F
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 22 (UZH2) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-2,5-bis(fluoranyl)phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O2E
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 21 (ADO_AD_089) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-3-fluoranyl-phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O09
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 7 (ADO_AC_074) | | Descriptor: | 6-[4-[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]-1-oxa-4,9-diazaspiro[5.5]undecan-9-yl]-N-(phenylmethyl)pyrimidin-4-amine, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O08
 
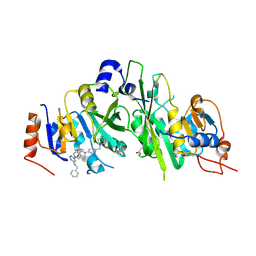 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 5 (ADO_AB_075) | | Descriptor: | 4-[[[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]amino]methyl]-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-4-ol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O0L
 
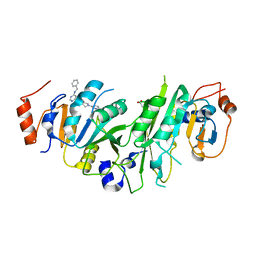 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 8 (ADO_AC_093) | | Descriptor: | 4-[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]-9-[6-[(phenylmethyl)amino]pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O29
 
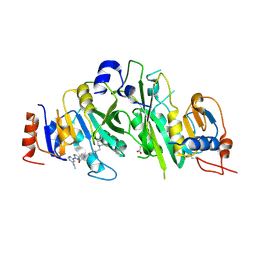 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 20 (ADO_AD_044) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-fluoranyl-phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7O2D
 
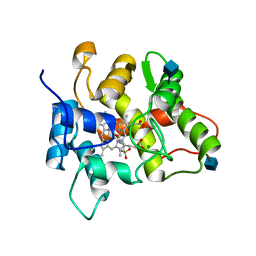 | |
7NZQ
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7O07
 
 | |
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7OJR
 
 | |
7OJS
 
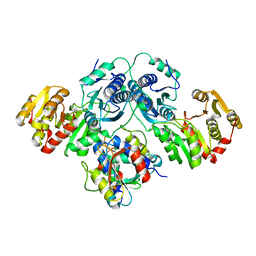 | |
6CHR
 
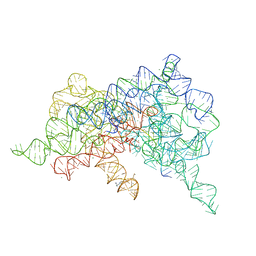 | | Crystal structure of a group II intron lariat with an intact 3' splice site (pre-2s state) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*A)-3'), RNA (621-MER), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
7NY7
 
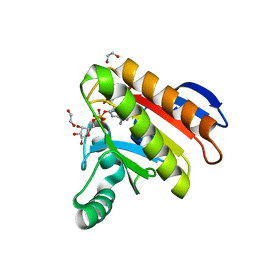 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OBM
 
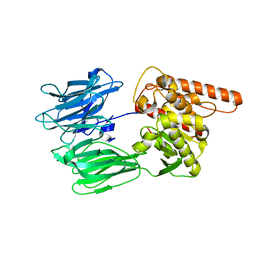 | | Crystal structure of the human Prolyl Endopeptidase-Like protein short form (residues 90-727) | | Descriptor: | Prolyl endopeptidase-like | | Authors: | Rosier, K, McDevitt, M.T, Brendan, J.F, Marcaida, M.J, Bingman, C.A, Pagliarini, D.J, Creemers, J.W.M, Smith, R.W, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prolyl endopeptidase-like is a (thio)esterase involved in mitochondrial respiratory chain function.
Iscience, 24, 2021
|
|
7NTL
 
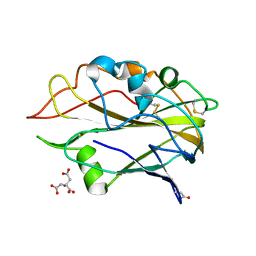 | | AA9 lytic polysaccharide monooxygenase (LPMO) from Malbranchea cinnamomea | | Descriptor: | CITRIC ACID, COPPER (II) ION, LPMO9F | | Authors: | Mazurkewich, S, Seveso, A, Huttner, S, Branden, G, Larsbrink, L. | | Deposit date: | 2021-03-10 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of a C1/C4-oxidizing AA9 lytic polysaccharide monooxygenase from the thermophilic fungus Malbranchea cinnamomea.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6CK4
 
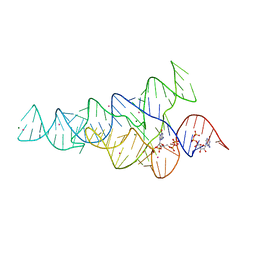 | | G96A mutant of the PRPP riboswitch from T. mathranii bound to ppGpp | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Reiss, C.W, Knappenberger, A.J, Strobel, S.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
7O1X
 
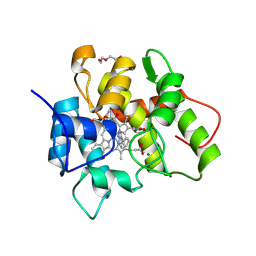 | |
7O2G
 
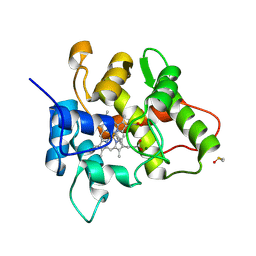 | |
7O1R
 
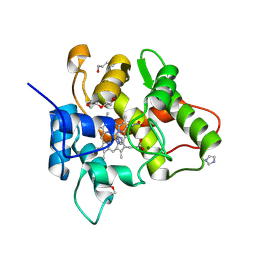 | |
7O1Z
 
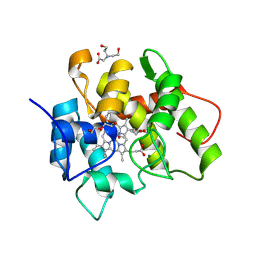 | |
7NYD
 
 | | cryoEM structure of 2C9-sMAC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Menny, A, Couves, E.C, Bubeck, D. | | Deposit date: | 2021-03-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
7ODF
 
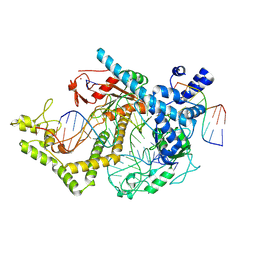 | | Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3 | | Descriptor: | Cas_phi3, DNA (5'-D(P*GP*G)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*TP*CP*AP*G)-3'), ... | | Authors: | Carabias del Rey, A, Fugilsang, A, Temperini, P, Pape, T, Sofos, N, Stella, S, Erledsson, S, Montoya, G. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3.
Nat Commun, 12, 2021
|
|
