6Z0R
 
 | | Crystal structure of SAICAR Synthetase (PurC) from Mycobacterium abscessus in complex with fragment 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanylpyrimidine-5-carbonitrile, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Thomas, S.E, Charoensutthivarakul, S, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.308 Å) | | Cite: | Development of Inhibitors of SAICAR Synthetase (PurC) from Mycobacterium abscessus Using a Fragment-Based Approach.
Acs Infect Dis., 8, 2022
|
|
6S36
 
 | | Crystal structure of E. coli Adenylate kinase R119K mutant | | Descriptor: | Adenylate kinase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2019-06-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleation of an Activating Conformational Change by a Cation-pi Interaction.
Biochemistry, 58, 2019
|
|
1ZY3
 
 | |
5JRD
 
 | | E. coli Hydrogenase-1 variant P508A | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Armstrong, F.A, Evans, R.M, Brooke, E.J, Islam, S.T.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Biochemistry, 56, 2017
|
|
1ZZK
 
 | | Crystal Structure of the third KH domain of hnRNP K at 0.95A resolution | | Descriptor: | Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
5MQY
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile, Cathepsin L1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors.
J. Med. Chem., 60, 2017
|
|
5MR0
 
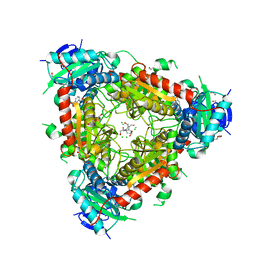 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
6UGF
 
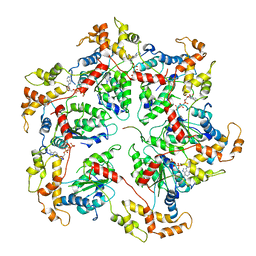 | |
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
6XWG
 
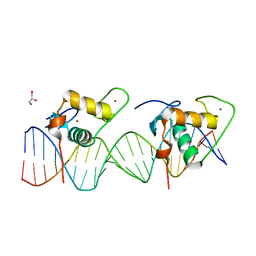 | | Crystal Structure of the Human RXR/RAR DNA-Binding Domain Heterodimer Bound to the Human RARb2 DR5 Response Element | | Descriptor: | CHLORIDE ION, GLYCEROL, RARb2 DR5 Response Element, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Peluso-Iltis, C, Rochel, N. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR-RXR.
Nucleic Acids Res., 48, 2020
|
|
6S07
 
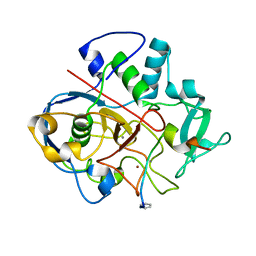 | | Structure of formylglycine-generating enzyme at 1.04 A in complex with copper and substrate reveals an acidic pocket for binding and acti-vation of molecular oxygen. | | Descriptor: | Abz-ALA-THR-THR-PRO-LEU-CYS-GLY-PRO-SER-ARG-ALA-SER-ILE-LEU-SER-GLY-ARG, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Leisinger, F, Miarzlou, D.A, Seebeck, F.P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of formylglycine-generating enzyme in complex with copper and a substrate reveals an acidic pocket for binding and activation of molecular oxygen.
Chem Sci, 10, 2019
|
|
5K67
 
 | | Designed Artificial Cupredoxins | | Descriptor: | GLYCEROL, Streptavidin, [CuII(biot-pr-dpea)]2+ | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Artificial Cupredoxins.
J.Am.Chem.Soc., 138, 2016
|
|
8QB1
 
 | | C-terminal domain of mirolase from Tannerella forsythia | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Mirolase, ... | | Authors: | Gomis-Ruth, F.X, Rodriguez-Banqueri, A, Mizgalska, D, Veillard, F, Goulas, T, Eckhard, U, Potempa, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system.
Open Biology, 14, 2024
|
|
7MQN
 
 | |
3K0W
 
 | | Crystal structure of the tandem IG-like C2-type 2 domains of the human mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Descriptor: | CHLORIDE ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, isoform 2 | | Authors: | Walker, J.R, Qiu, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tandem Ig-Like C2-Type 2 Domains of the Human Mucosa-Associated Lymphoid Tissue Lymphoma Translocation Protein 1.
To be Published
|
|
1HEK
 
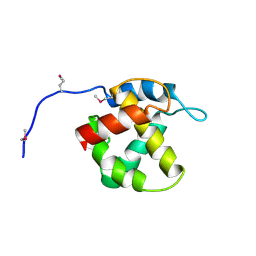 | | Crystal structure of equine infectious anaemia virus matrix antigen (EIAV MA) | | Descriptor: | GAG POLYPROTEIN, CORE PROTEIN P15 | | Authors: | Hatanaka, H, Iourin, O, Rao, Z, Fry, E, Kingsman, A, Stuart, D.I. | | Deposit date: | 2000-11-24 | | Release date: | 2001-11-23 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Equine Infectious Anemia Virus Matrix Protein.
J.Virol., 76, 2002
|
|
2OXS
 
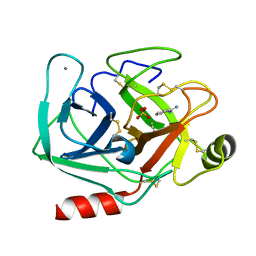 | | Crystal Structure of the trypsin complex with benzamidine at high temperature (35 C) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Alok, A, Sinha, M, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the trypsin complex with benzamidine at high temperature (35 C)
To be Published
|
|
5JT7
 
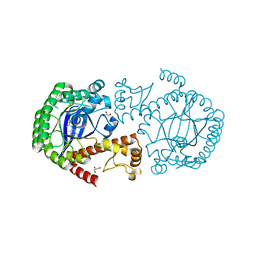 | |
6P5Q
 
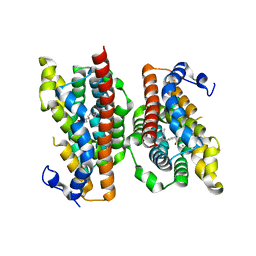 | | X-ray structure of Fe(II)-soaked UndA bound to lauric acid | | Descriptor: | FE (III) ION, GLYCEROL, LAURIC ACID, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate-Triggered Formation of a Peroxo-Fe2(III/III) Intermediate during Fatty Acid Decarboxylation by UndA.
J.Am.Chem.Soc., 141, 2019
|
|
6I4V
 
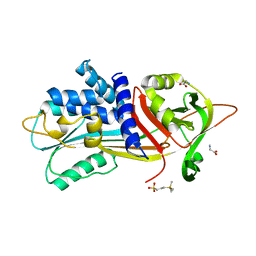 | |
6S7M
 
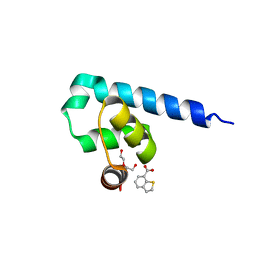 | |
7QF8
 
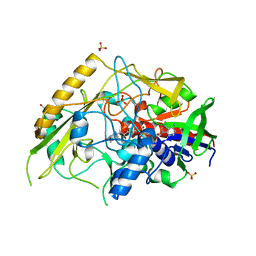 | | Crystal structure of a bacterial pyranose 2-oxidase from Pseudoarthrobacter siccitolerans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, GMC oxidoreductase family protein, ... | | Authors: | Borges, P.T, Frazao, T, Taborda, A, Frazao, C, Martins, L.O. | | Deposit date: | 2021-12-04 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Mechanistic insights into glycoside 3-oxidases involved in C-glycoside metabolism in soil microorganisms.
Nat Commun, 14, 2023
|
|
6P1J
 
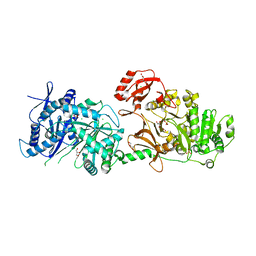 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo2 serine module | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-20 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
8E9D
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS bound to maleic acid at 100 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
4P7A
 
 | | Crystal Structure of human MLH1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Mlh1, MAGNESIUM ION, ... | | Authors: | Tempel, W, Lam, R, Zeng, H, Walker, J.R, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human MLH1 N-terminus: implications for predisposition to Lynch syndrome.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
