6TYN
 
 | | Salmonella Typhi PltB Homopentamer with Neu-5NAc-9OAc-alpha-2-3-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertussis like toxin subunit B | | Authors: | Nguyen, T, Milano, S.K, Yang, Y.A, Song, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication.
Plos Pathog., 16, 2020
|
|
4YFV
 
 | | X-ray structure of the 4-N-formyltransferase VioF from Providencia alcalifaciens O30 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, VioF | | Authors: | Genthe, N.A, Thoden, J.B, Benning, M.M, Holden, H.M. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular structure of an N-formyltransferase from Providencia alcalifaciens O30.
Protein Sci., 24, 2015
|
|
1AGT
 
 | | SOLUTION STRUCTURE OF THE POTASSIUM CHANNEL INHIBITOR AGITOXIN 2: CALIPER FOR PROBING CHANNEL GEOMETRY | | Descriptor: | AGITOXIN 2 | | Authors: | Krezel, A.M, Kasibhatla, C, Hidalgo, P, Mackinnon, R, Wagner, G. | | Deposit date: | 1995-04-14 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the potassium channel inhibitor agitoxin 2: caliper for probing channel geometry.
Protein Sci., 4, 1995
|
|
8ACT
 
 | | structure of the human beta-cardiac myosin folded-back off state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light chain 3, ... | | Authors: | Grinzato, A, Kandiah, E, Robert-Paganin, J, Auguin, D, Kikuti, C, Nandwani, N, Moussaoui, D, Pathak, D, Ruppel, K.M, Spudich, J.A, Houdusse, A. | | Deposit date: | 2022-07-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the folded-back state of human beta-cardiac myosin.
Nat Commun, 14, 2023
|
|
6I7H
 
 | | Crystal structure of dimeric FICD mutant K256S | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6S49
 
 | |
5M75
 
 | |
7APM
 
 | | tRNA-guanine transglycosylase H319C mutant spin-labeled with MTSL. | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-10-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unraveling a Ligand-Induced Twist of a Homodimeric Enzyme by Pulsed Electron-Electron Double Resonance.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6PJI
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-43 | | Descriptor: | Protease NL4-3, SULFATE ION, methyl [(1S)-1-cyclopropyl-2-({(2S,3S,5S)-5-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-1,6-diphenylhexan-2-yl}amino)-2-oxoethyl]carbamate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
4ECD
 
 | | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase | | Descriptor: | CHLORIDE ION, Chorismate synthase | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase
TO BE PUBLISHED
|
|
8DK2
 
 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetABC in an unclamped state trapped in ATP dependent dimeric form | | Descriptor: | DNA (26-MER), JetA, JetB, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
4YH7
 
 | | Crystal structure of PTPdelta ectodomain in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
7R14
 
 | | Alpha Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
5MAB
 
 | | FoxE P3121 crystal structure of Rhodopseudomonas ferrooxidans SW2 putative iron oxidase | | Descriptor: | COPPER (II) ION, FoxE, HEME C, ... | | Authors: | Pereira, L, Saraiva, I.H, Oliveira, A.S, Soares, C, Gomes, R.O, Frazao, C. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | FoxE P3121 crystal structure of Rhodopseudomonas ferrooxidans SW2 putative iron oxidase
To be published
|
|
5J18
 
 | | Solution structure of Ras Binding Domain (RBD) of B-Raf complexed with Rigosertib (Complex I) | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
6S6T
 
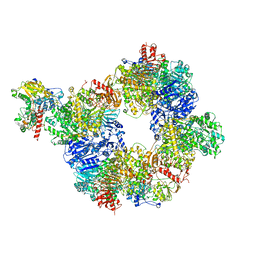 | |
8G1U
 
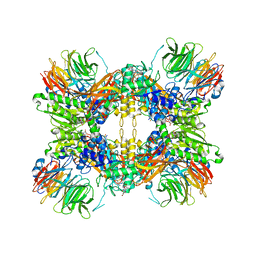 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
7B2X
 
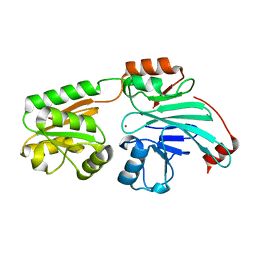 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
5M60
 
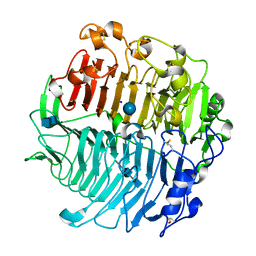 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, SODIUM ION, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
4RDM
 
 | | Crystal structure of R.NgoAVII restriction endonuclease B3 domain with cognate DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*AP*GP*CP*GP*GP*CP*AP*AP*TP*CP*C)-3)', DNA (5'-D(*GP*GP*GP*AP*TP*TP*GP*CP*CP*GP*CP*TP*TP*AP*G)-3)', Restriction endonuclease R.NgoVII, ... | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
8GF2
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies eCR3022.20 and CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Broadening a SARS-CoV-1-neutralizing antibody for potent SARS-CoV-2 neutralization through directed evolution.
Sci.Signal., 16, 2023
|
|
2N5Q
 
 | | Solution structure of cystein-rich peptide jS1 from Jasminum sambac | | Descriptor: | cysteine-rich peptide jS1 | | Authors: | Shin, J, Kumari, G, Serra, A, Nguyen, P.Q.T, Yoon, H, Tam, J.P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification of a cysteine-rich peptide family with unusual disulfide connectivity from Jasminum sambac
To be Published
|
|
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4LE5
 
 | |
