7ZYW
 
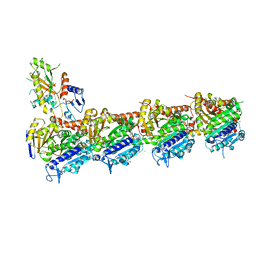 | | Crystal structure of T2R-TTL-PM534 complex | | Descriptor: | (4R)-N-[(1R)-1-[4-(cyclopropylmethoxy)-6-oxidanylidene-pyran-2-yl]butyl]-4-methyl-2-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]-5H-1,3-thiazole-4-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Diaz, J.F, Cuevas, C. | | Deposit date: | 2022-05-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | PM534, an Optimized Target-Protein Interaction Strategy through the Colchicine Site of Tubulin.
J.Med.Chem., 67, 2024
|
|
4U07
 
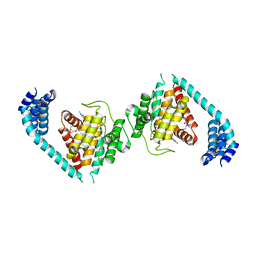 | | ATP bound to eukaryotic FIC domain containing protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
6SIV
 
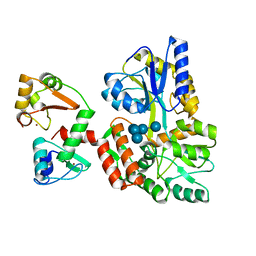 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-08-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
7NYC
 
 | | cryoEM structure of 3C9-sMAC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Menny, A, Couves, E.C, Bubeck, D. | | Deposit date: | 2021-03-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
6MT8
 
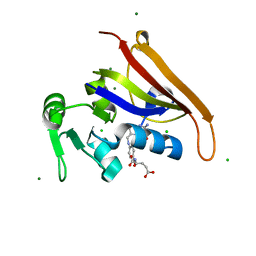 | | E. coli DHFR complex modeled with two ligand states | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, DIHYDROFOLIC ACID, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate.
Struct Dyn., 6, 2019
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6G3S
 
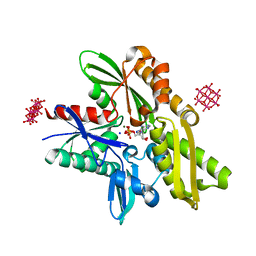 | | Structure of tellurium-centred Anderson-Evans polyoxotungstate (TEW) bound to the nucleotide binding domain of HSP70. Second structure of two TEW-HSP70 structures deposited. | | Descriptor: | 6-tungstotellurate(VI), ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1A, ... | | Authors: | Mac Sweeney, A, Chambovey, A, Wicki, M, Mueller, M, Artico, N, Lange, R, Bijelic, A, Breibeck, J, Rompel, A. | | Deposit date: | 2018-03-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystallization additive hexatungstotellurate promotes the crystallization of the HSP70 nucleotide binding domain into two different crystal forms.
PLoS ONE, 13, 2018
|
|
6SGS
 
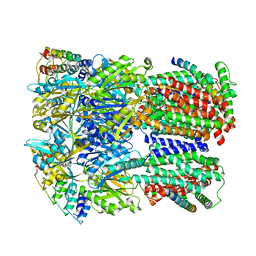 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc | | Descriptor: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
8OQX
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK with a phosphate analogue | | Descriptor: | 1,2-ETHANEDIOL, ATPase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
6N3H
 
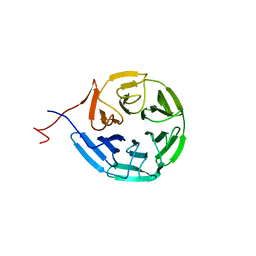 | | Crystal structure of Kelch domain of the human NS1 binding protein | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Zhang, K, Shang, G, Padavannil, A, Fontoura, B.M.A, Chook, Y.M. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4Y3T
 
 | | Endothiapepsin in complex with fragment 207 | | Descriptor: | (3R)-piperidin-3-yl(piperidin-1-yl)methanone, (3S)-piperidin-3-yl(piperidin-1-yl)methanone, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Huschmann, F.U, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
7L7K
 
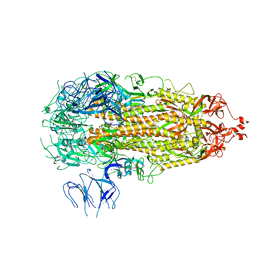 | |
1AJ6
 
 | |
6GA9
 
 | | BACTERIORHODOPSIN, 390 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
5JML
 
 | | X-ray structure of the complex between bovine pancreatic ribonuclease and penthachlorocarbonyliridate(III) (2 months of soaking) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, IRIDIUM ION, ... | | Authors: | Caterino, M, Petruk, A.A, Vergara, A, Ferraro, G, Merlino, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mapping the protein-binding sites for iridium(iii)-based CO-releasing molecules.
Dalton Trans, 45, 2016
|
|
4Y5A
 
 | | Endothiapepsin in complex with fragment 206 | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-(trifluoromethyl)furan-2-yl]methanamine, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Huschmann, F.U, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
6ZEK
 
 | | Crystal structure of mouse CSAD | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Cysteine sulfinic acid decarboxylase, ... | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J.Struct.Biol., 213, 2021
|
|
4Y76
 
 | | Factor Xa complex with GTC000401 | | Descriptor: | CALCIUM ION, Coagulation factor X, N~2~-[(6-chloronaphthalen-2-yl)sulfonyl]-N~2~-{(3S)-1-[(2S)-1-(4-methyl-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}glycinamide | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4Y79
 
 | | Factor Xa complex with GTC000406 | | Descriptor: | (E)-2-(4-chlorophenyl)-N-{(3S)-1-[(2S)-1-(morpholin-4-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}ethenesulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
8QB1
 
 | | C-terminal domain of mirolase from Tannerella forsythia | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Mirolase, ... | | Authors: | Gomis-Ruth, F.X, Rodriguez-Banqueri, A, Mizgalska, D, Veillard, F, Goulas, T, Eckhard, U, Potempa, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system.
Open Biology, 14, 2024
|
|
5O7A
 
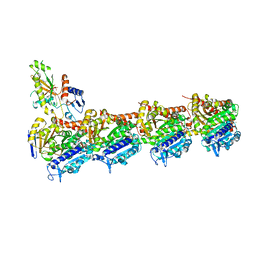 | | Quinolin-6-yloxyacetamides are microtubule destabilizing agents that bind to the colchicine site of tubulin | | Descriptor: | (2~{R})-2-(3-ethynylquinolin-6-yl)oxy-2-methoxy-~{N}-[(1~{E})-1-methoxyimino-2-methyl-propan-2-yl]ethanamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, A, Olieric, N, Steinmetz, M.O. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Quinolin-6-Yloxyacetamides Are Microtubule Destabilizing Agents That Bind to the Colchicine Site of Tubulin.
Int J Mol Sci, 18, 2017
|
|
6G7K
 
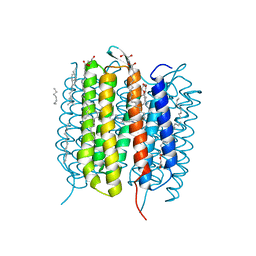 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 10 ps state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
4Y7F
 
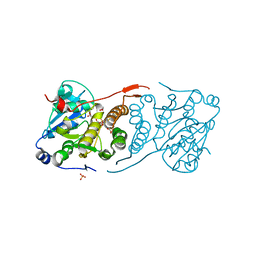 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and 3-(phosphonooxy)propanoic acid (PPA) - GpgS Mn2+ UDP-Glc PPA | | Descriptor: | 1,2-ETHANEDIOL, 3-(phosphonooxy)propanoic acid, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.231 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6CW2
 
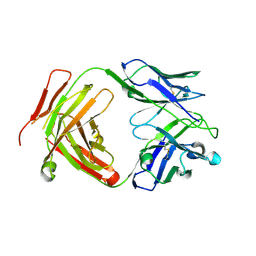 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 1 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4Y7G
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and glycerol 3-phosphate (G3P) - GpgS Mn2+ UDP-Glc G3P | | Descriptor: | Glucosyl-3-phosphoglycerate synthase, MANGANESE (II) ION, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
