7AMZ
 
 | | Crystal structure of human Butyrylcholinesterase in complex with N-((2S,3R)-4-((2,2-dimethylpropyl)amino)-3-hydroxy-1-phenylbutan-2-yl)-2,2-diphenylacetamide | | Descriptor: | 2,2-dimethylpropyl-[(2~{R},3~{S})-3-(2,2-diphenylethanoylamino)-2-oxidanyl-4-phenyl-butyl]azanium, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Pasieka, A, Panek, D, Wieckowska, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of multifunctional anti-Alzheimer's agents with a unique mechanism of action including inhibition of the enzyme butyrylcholinesterase and gamma-aminobutyric acid transporters.
Eur.J.Med.Chem., 218, 2021
|
|
6PGQ
 
 | |
5MUN
 
 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
6SS4
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
7TBJ
 
 | | Composite structure of the human nuclear pore complex (NPC) symmetric core generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | NUP107 CTD, NUP107 NTD, NUP133, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
6PH2
 
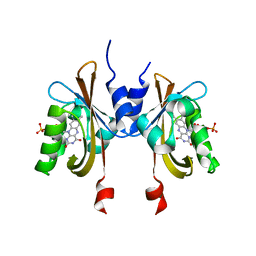 | | Complete LOV domain from the LOV-HK sensory protein from Brucella abortus (mutant C69S, construct 15-155) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
7QIK
 
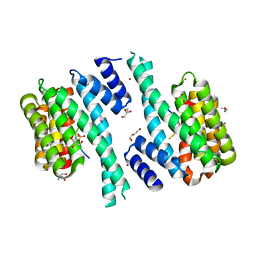 | | SARS-CoV-2 Nucleocapsid phosphopeptide 193-200 bound to human 14-3-3 sigma | | Descriptor: | 14-3-3 protein sigma, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Smith, J.L.R, Antson, A.A. | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | SARS-CoV-2 Nucleocapsid phosphopeptide 193-200 bound to human 14-3-3 sigma
To Be Published
|
|
6YRO
 
 | | Streptococcus suis SadP mutant - N285D | | Descriptor: | GLYCEROL, SODIUM ION, SadP | | Authors: | Papageorgiou, A.C, Haataja, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The binding mechanism of the virulence factor Streptococcus suis adhesin P subtype to globotetraosylceramide is associated with systemic disease.
J.Biol.Chem., 295, 2020
|
|
6SC2
 
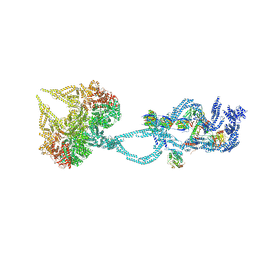 | | Structure of the dynein-2 complex; IFT-train bound model | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Toropova, K, Zalyte, R, Mukhopadhyay, A.G, Mladenov, M, Carter, A.P, Roberts, A.J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6V71
 
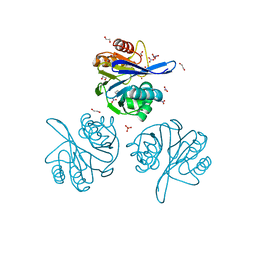 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
7QIP
 
 | |
6MJT
 
 | | Azurin 122F/124W/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
6V7I
 
 | | Structure of KPC-2 bound to Vaborbactam at 1.25 A | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, SULFATE ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-12-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Binding Kinetics of Vaborbactam in Class A beta-Lactamase Inhibition.
Antimicrob.Agents Chemother., 64, 2020
|
|
6MIA
 
 | | Crystal structure of CTX-M-14 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Beta-lactamase | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
7AIP
 
 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Liko, I, Tehan, B.G, Almeida, F.G, Elkins, J, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
6V8K
 
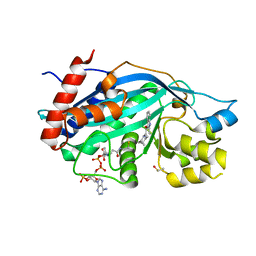 | |
6MJP
 
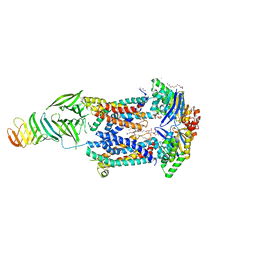 | | LptB(E163Q)FGC from Vibrio cholerae | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 6-cyclohexylhexyl beta-D-glucopyranoside, ABC transporter ATP-binding protein, ... | | Authors: | Owens, T.W, Kahne, D, Kruse, A.C. | | Deposit date: | 2018-09-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of unidirectional export of lipopolysaccharide to the cell surface.
Nature, 567, 2019
|
|
6PNM
 
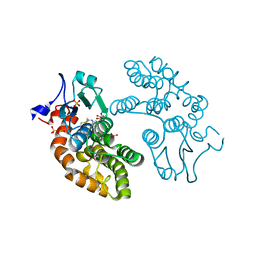 | |
6VCQ
 
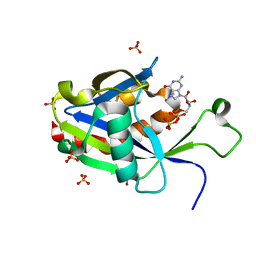 | | Crystal structure of E.coli RppH in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
5M75
 
 | |
7AK7
 
 | | Structure of Salmonella TacT2 toxin bound to TacA2 antitoxin | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CHLORIDE ION, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7O49
 
 | |
6YIJ
 
 | | Crystal structure of the CREBBP bromodomain in complex with a benzo-diazepine ligand | | Descriptor: | (4~{R})-6-[(~{E})-5-(7-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)pent-1-enyl]-4-methyl-1,3,4,5-tetrahydro-1,5-benzodiazepin-2-one, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a benzo-diazepine ligand
To Be Published
|
|
7SPN
 
 | | Crystal structure of IS11, a thermophilic esterase | | Descriptor: | IS11 | | Authors: | Stogios, P.J, Evdokimova, E, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of IS11, a thermophilic esterase
To Be Published
|
|
6VDW
 
 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Manganese(III) Porphyrin and Substrate 4-nitrophenol | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
