8EH5
 
 | | Cryo-EM structure of L9 Fab in complex with rsCSP | | Descriptor: | Circumsporozoite protein, L9 Heavy chain, L9 Light chain | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-09-13 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of epitope selectivity and potent protection from malaria by PfCSP antibody L9.
Nat Commun, 14, 2023
|
|
8CRT
 
 | | Local refinement of Rh trimer, glycophorin B and Band3-III transmembrane region, class 1a of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ammonium transporter Rh type A, Band 3 anion transport protein, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PXD
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state B) | | Descriptor: | AAA ATPase forming ring-shaped complexes, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
6FAI
 
 | | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Scaiola, A, Pena, C, Weisser, M, Boehringer, D, Leibundgut, M, Klingauf-Nerurkar, P, Gerhardy, S, Panse, V.G, Ban, N. | | Deposit date: | 2017-12-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit.
EMBO J., 37, 2018
|
|
7PFS
 
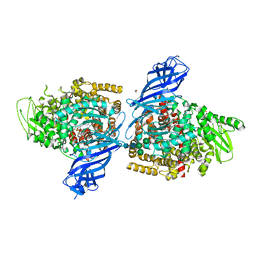 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
6OLA
 
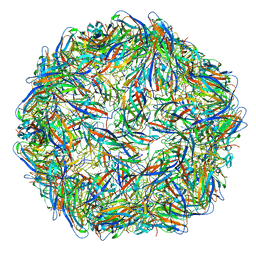 | | Structure of the PCV2d virus-like particle | | Descriptor: | Capsid protein, DNA (5'-D(P*CP*CP*GP*G)-3') | | Authors: | Khayat, R, Wen, K, Alimova, A, Galarza, J, Gottlieb, P. | | Deposit date: | 2019-04-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural characterization of the PCV2d virus-like particle at 3.3 angstrom resolution reveals differences to PCV2a and PCV2b capsids, a tetranucleotide, and an N-terminus near the icosahedral 3-fold axes.
Virology, 537, 2019
|
|
7B32
 
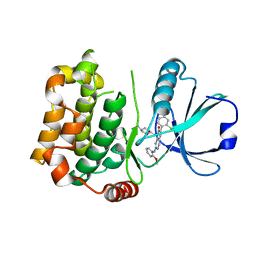 | | MST3 in complex with MRIA7 | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-[(2-methoxyphenyl)amino]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8C13
 
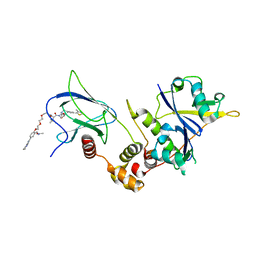 | | Crystal structure of pVHL:ElonginC:ElonginB complex bound to PROTAC JW48 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[3-[2-[2-[2-(acetamidomethyl)-4-(6,7-dihydro-5~{H}-pyrrolo[1,2-a]imidazol-2-yl)phenoxy]ethoxy]ethoxy]propanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Weckesser, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
6OLO
 
 | |
8EKO
 
 | |
6X9Z
 
 | |
8C7K
 
 | |
6OWO
 
 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 CORE BOUND TO NECAP | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J.S, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-09-11 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
6TUY
 
 | | Human LSD1/CoREST bound to the quinazoline inhibitor MC4106 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mattevi, A, Marrocco, B. | | Deposit date: | 2020-01-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel non-covalent LSD1 inhibitors endowed with anticancer effects in leukemia and solid tumor cellular models.
Eur.J.Med.Chem., 237, 2022
|
|
6TXJ
 
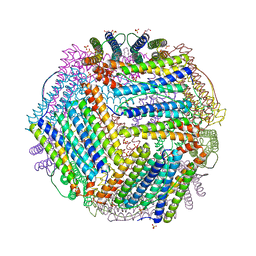 | | Crystal structure of thermotoga maritima A42V E65D Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S, Biela, A.P. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
5LUD
 
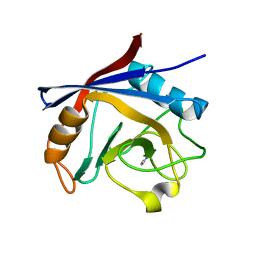 | | Structure of Cyclophilin A in complex with 2,3-Diaminopyridine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, pyridine-2,3-diamine | | Authors: | McNae, I.W, Nowicki, M.W, Blackburn, E.A, Wear, M.A, Walkinshaw, M.D. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermo-kinetic analysis space expansion for cyclophilin-ligand interactions - identification of a new nonpeptide inhibitor using BiacoreTM T200.
FEBS Open Bio, 7, 2017
|
|
5J54
 
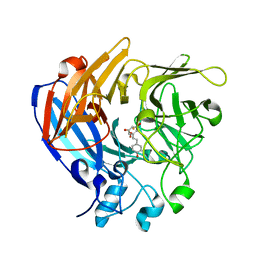 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8C7Y
 
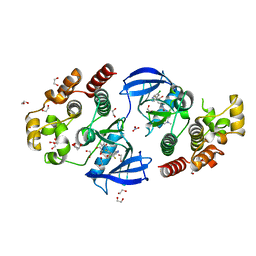 | | Crystal structure of BRAF V600E in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
8C7X
 
 | | Crystal structure of BRAF in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
8CLM
 
 | |
6OR4
 
 | | Crystal structure of SpGH29 | | Descriptor: | Glycoside hydrolase, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-29 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
6XJ7
 
 | |
6XJ6
 
 | |
6O7P
 
 | | Nitrogenase MoFeP mutant F99Y from Azotobacter vinelandii in the dithionite reduced state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
5J79
 
 | | The identification and pharmacological characterization of 6-(tert-butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a highly potent and selective inhibitor of RIP2 Kinase, Compound 3 complex | | Descriptor: | 4-methyl-3-{[6-(methylsulfonyl)quinolin-4-yl]amino}phenol, Receptor-interacting serine/threonine-protein kinase 2, SULFATE ION | | Authors: | Convery, M.A, Casillas, L.N, Haile, P.A, Votta, B.J, Lakdawala, A.S. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The Identification and Pharmacological Characterization of 6-(tert-Butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a Highly Potent and Selective Inhibitor of RIP2 Kinase.
J.Med.Chem., 59, 2016
|
|
