7T9L
 
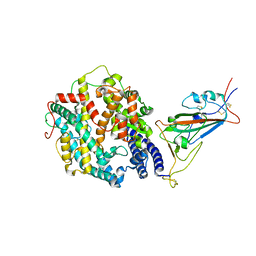 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
2OSZ
 
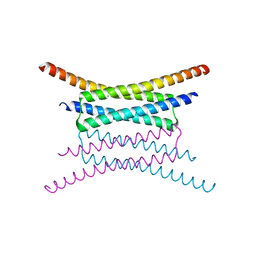 | |
5XNX
 
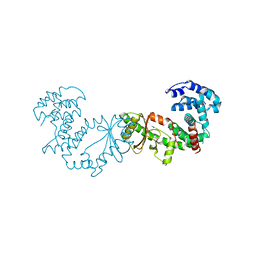 | | Crystallographic structure of the enzymatically active N-terminal domain of the Rel protein from Mycobacterium tuberculosis | | Descriptor: | Bifunctional (p)ppGpp synthase/hydrolase RelA, MAGNESIUM ION | | Authors: | Singal, B, Balakrishna, A.M, Manimekalai, M.S.S, Nartey, W, Gruber, G. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystallographic and solution structure of the N-terminal domain of the Rel protein from Mycobacterium tuberculosis
FEBS Lett., 591, 2017
|
|
4DRO
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH (1R)-3-(3,4-dimethoxyphenyl)-1-phenylpropyl (2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
2OPI
 
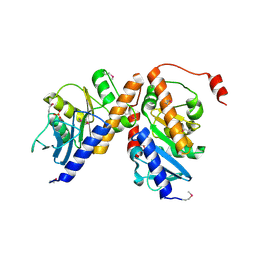 | |
4DRZ
 
 | | Crystal structure of human RAB14 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-14 | | Authors: | Wang, J, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-17 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human RAB14
to be published
|
|
1TTF
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
2P3N
 
 | | Thermotoga maritima IMPase TM1415 | | Descriptor: | Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Stieglitz, K.A, Roberts, M.F, Li, W, Stec, B. | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J., 274, 2007
|
|
4DF7
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23L/V99I at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
2OTG
 
 | | Rigor-like structures of muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2007-02-08 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Rigor-like Structures from Muscle Myosins Reveal Key Mechanical Elements in the Transduction Pathways of This Allosteric Motor.
Structure, 15, 2007
|
|
6EU3
 
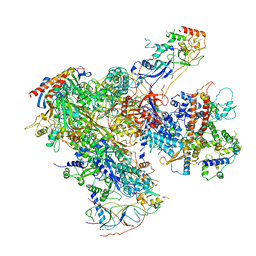 | | Apo RNA Polymerase III - closed conformation (cPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
2P7T
 
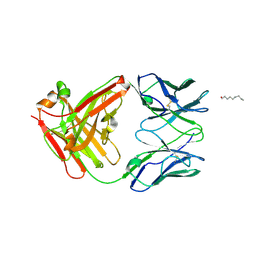 | | Crystal Structure of KcsA mutant | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, FAB-A, FAB-B, ... | | Authors: | Cordero-Morales, J.F, Vishwanath, J, Lewis, A, Valeria, V.R, Cortes, D.M, Roux, B, Perozo, E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-10-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of KcsA mutant
To be Published
|
|
6F8N
 
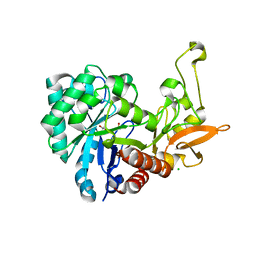 | | Key residues affecting transglycosylation activity in family 18 chitinases - Insights into donor and acceptor subsites | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Madhuprakash, J, Dalhus, B, Swaroopa Rani, T, Podile, A.R, Eijsink, V.G.H, Sorlie, M. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Key Residues Affecting Transglycosylation Activity in Family 18 Chitinases: Insights into Donor and Acceptor Subsites.
Biochemistry, 57, 2018
|
|
2OX6
 
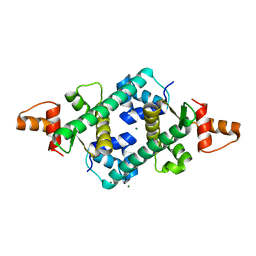 | |
4LQ2
 
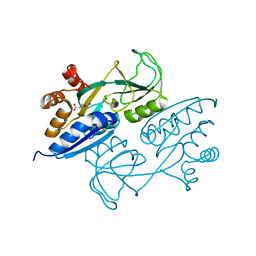 | | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium in complex with effector ligand, O-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, O-ACETYLSERINE | | Authors: | Mittal, M, Singh, A.K, Kumaran, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Crystal structure of ligand binding domain of CysB, a LysR member from Salmonella typhimurium in complex with effector ligand, O-acetylserine
TO BE PUBLISHED
|
|
3NNC
 
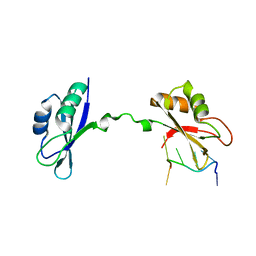 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2005 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
4DTZ
 
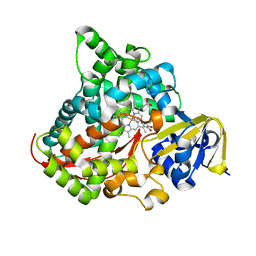 | | cytochrome P450 BM3h-8C8 MRI sensor bound to dopamine | | Descriptor: | L-DOPAMINE, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 8C8 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4DUH
 
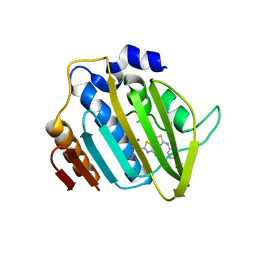 | | Crystal structure of 24 kDa domain of E. coli DNA gyrase B in complex with small molecule inhibitor | | Descriptor: | 4-{[4'-methyl-2'-(propanoylamino)-4,5'-bi-1,3-thiazol-2-yl]amino}benzoic acid, DNA gyrase subunit B | | Authors: | Brvar, M, Renko, M, Perdih, A, Solmajer, T, Turk, D. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of substituted 4,5'-bithiazoles as novel DNA gyrase inhibitors.
J.Med.Chem., 55, 2012
|
|
4DUP
 
 | | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | quinone oxidoreductase | | Authors: | Kumaran, D, Rice, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
7T8W
 
 | | Structure of antibody 3G12 bound to Respiratory Syncytial Virus G central conserved domain mutant S177Q | | Descriptor: | 3G12 Fab Heavy chain, 3G12 Fab Light chain, Mature secreted glycoprotein G | | Authors: | Nunez Castrejon, A.M, O'Rourke, S.M, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design and Antigenic Validation of Respiratory Syncytial Virus G Immunogens.
J.Virol., 96, 2022
|
|
2PBP
 
 | | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426 | | Descriptor: | Enoyl-CoA hydratase subunit I | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki, R.C, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426
To be Published
|
|
4M05
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii R173E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chlorite dismutase, ... | | Authors: | Gysel, K, Hagmueller, A, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
5XO2
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide II | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2,4-dideoxy-alpha-D-xylo-hexopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Envelope glycoprotein B | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, S, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-25 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
6F08
 
 | | 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1) | | Descriptor: | 14-3-3 protein zeta/delta, PENTAETHYLENE GLYCOL, Son of sevenless homolog 1 | | Authors: | Ballone, A, Centorrino, F, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1).
J. Struct. Biol., 202, 2018
|
|
3O6C
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
