8GZ5
 
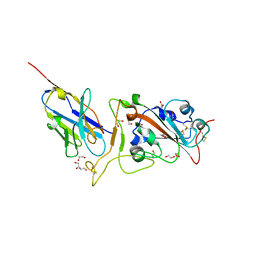 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P17, ... | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
6O5V
 
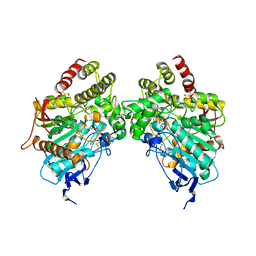 | | Binary complex of native hAChE with oxime reactivator RS-170B | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase.
J.Biol.Chem., 294, 2019
|
|
6YQO
 
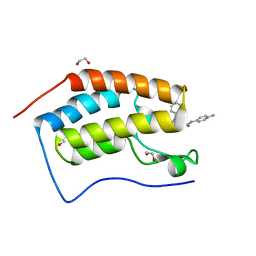 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW12 | | Descriptor: | (S)-N1-(4-(2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetamido)phenyl)-N8-hydroxyoctanediamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
6QDK
 
 | |
6UND
 
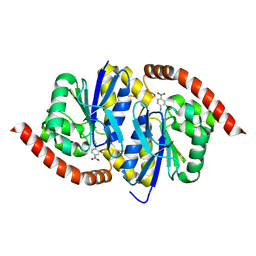 | |
6UE3
 
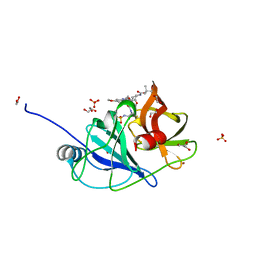 | |
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
7OML
 
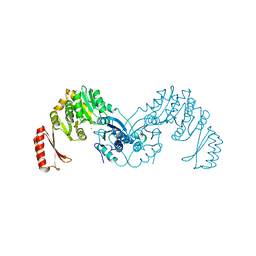 | |
8ECY
 
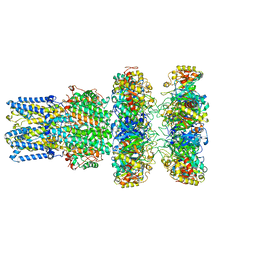 | | cryoEM structure of bovine bestrophin-2 and glutamine synthetase complex | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Owji, A.P, Kittredge, A.K, Yang, T. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Bestrophin-2 and glutamine synthetase form a complex for glutamate release.
Nature, 611, 2022
|
|
7OLH
 
 | |
6YCY
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Krementsova, E.B, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Elife, 9, 2020
|
|
7BEU
 
 | | Human glutathione transferase M1-1 | | Descriptor: | Glutathione S-transferase Mu 1, SODIUM ION | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2020-12-28 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Ligandability Assessment of Human Glutathione Transferase M1-1 Using Pesticides as Chemical Probes.
Int J Mol Sci, 23, 2022
|
|
7BB0
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with NAT22-366511 and PKI (5-24) | | Descriptor: | (R)-3,3-dimethyl-5-oxo-5-(6-(3-(pyridin-4-yl)-1,2,4-oxadiazol-5-yl)-1,4,6,7-tetrahydro-5H-imidazo[4,5-c]pyridin-5-yl)pentanoic acid, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Oebbeke, M, Mueller, J, Glinca, S, Heine, A, Klebe, G. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with NAT22-366511 and PKI (5-24)
To Be Published
|
|
6UIZ
 
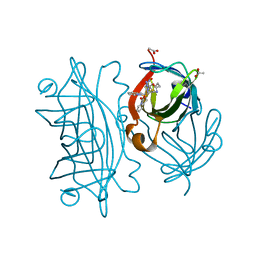 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(triaza-1,2-dien-2-ium-1-ide-kappaN~1~)iron(4+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
7BC1
 
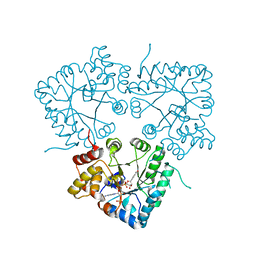 | |
6YR8
 
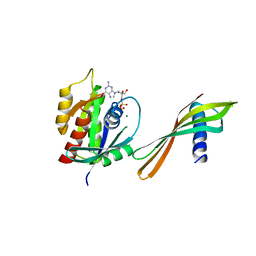 | | Affimer K6 - KRAS protein complex | | Descriptor: | Affimer K6, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
8GYH
 
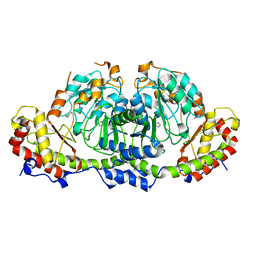 | | Crystal structure of Fic25 (apo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
6YAT
 
 | | Crystal structure of STK4 (MST1) in complex with compound 6 | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 4-[5-(3-chlorophenyl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]morpholine, GLYCEROL, ... | | Authors: | Chaikuad, A, Bata, N, Limpert, A.S, Lambert, L.J, Bakas, N.A, Cosford, N.D.P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Inhibitors of the Hippo Pathway Kinases STK3/MST2 and STK4/MST1 Have Utility for the Treatment of Acute Myeloid Leukemia.
J.Med.Chem., 65, 2022
|
|
8GOP
 
 | | SARS-CoV-2 specific private TCR RLQ7 | | Descriptor: | SARS-CoV-2 specific private TCR RLQ7 alpha, SARS-CoV-2 specific private TCR RLQ7 beta | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
8GOM
 
 | | SARS-CoV-2 specific private TCR RLQ7 in complex with RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 specific private TCR RLQ7 alpha, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
6ON5
 
 | |
6ON8
 
 | |
8HZD
 
 | | A new fluorescent RNA aptamer bound with N618 | | Descriptor: | 4-[(~{E})-2-[(4~{Z})-4-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-1-methyl-5-oxidanylidene-imidazol-2-yl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, RNA (36-MER) | | Authors: | Song, Q.Q, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZJ
 
 | | A new fluorescent RNA aptamer bound with N571 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8V4U
 
 | |
