4NCQ
 
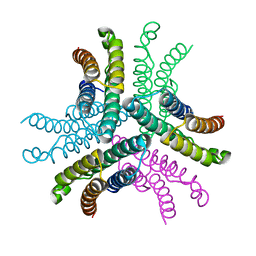 | | Crystal structure of NiSOD H1A mutant | | Descriptor: | Superoxide dismutase [Ni] | | Authors: | Guce, A.I, Garman, S.C. | | Deposit date: | 2013-10-24 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Nickel superoxide dismutase: structural and functional roles of His1 and its H-bonding network.
Biochemistry, 54, 2015
|
|
6D0H
 
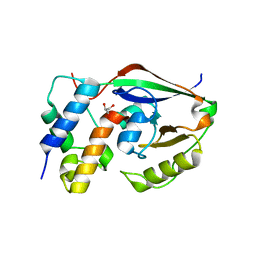 | |
6VJ0
 
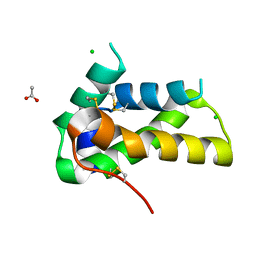 | | Crystal structure of a chitin-binding protein from Moringa oleifera seeds (Mo-CBP4) | | Descriptor: | ACETATE ION, CHLORIDE ION, Chitin-binding protein Mo-CBP4 | | Authors: | Bezerra, E.H.S, Lopes, T.D.P, da Silva, F.M.S, Costa, H.P.S, Freire, V.N, Rocha, B.A.M, Sousa, D.O.B. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a lectin from Moringa oleifera seeds with imflammatory activities
To Be Published
|
|
8CRB
 
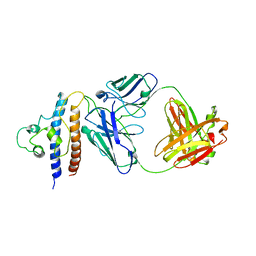 | | Cryo-EM structure of PcrV/Fab(11-E5) | | Descriptor: | Heavy chain, Light chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
5I3V
 
 | | Crystal structure of BACE1 in complex with aminoquinoline compound 1 | | Descriptor: | (2R)-3-[2-amino-6-(3-methylpyridin-2-yl)quinolin-3-yl]-N-(3,3-dimethylbutyl)-2-methylpropanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-Linking Approach Using (19)F NMR Spectroscopy To Obtain Highly Potent and Selective Inhibitors of beta-Secretase.
J.Med.Chem., 59, 2016
|
|
6D0X
 
 | | Crystal structure of chimeric H.2140 / K.1210 Fab in complex with circumsporozoite protein NANP3 | | Descriptor: | 1210 Antibody, light chain, 2140 Antibody, ... | | Authors: | Scally, S.W, Bosch, A, Imkeller, K, Wardemann, H, Julien, J.P. | | Deposit date: | 2018-04-11 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope.
Science, 360, 2018
|
|
8CR9
 
 | | Cryo-EM structure of PcrV/Fab(30-B8) | | Descriptor: | Heavy chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV, light chain | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
3U6J
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyrazolone inhibitor | | Descriptor: | N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3KDQ
 
 | | Crystal structure of a functionally unknown conserved protein from Corynebacterium diphtheriae. | | Descriptor: | uncharacterized conserved protein | | Authors: | Zhang, R, Wu, R, Tan, K, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a functionally unknown conserved protein from Corynebacterium diphtheriae.
To be Published
|
|
5URS
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P178 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P178
To Be Published
|
|
7UHM
 
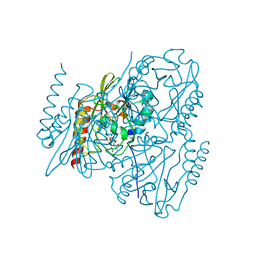 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (150 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
5OVW
 
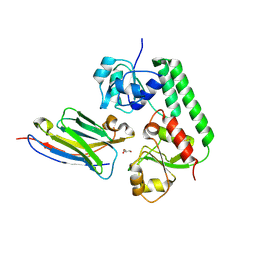 | | Nanobody-bound BtuF, the vitamin B12 binding protein in Escherichia coli | | Descriptor: | GLYCEROL, Nanobody, Vitamin B12-binding protein | | Authors: | Mireku, S.A, Sauer, M.M, Glockshuber, R, Locher, K.P. | | Deposit date: | 2017-08-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural basis of nanobody-mediated blocking of BtuF, the cognate substrate-binding protein of the Escherichia coli vitamin B12 transporter BtuCD.
Sci Rep, 7, 2017
|
|
5V7C
 
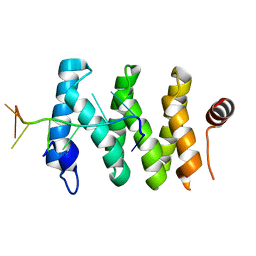 | |
4WW1
 
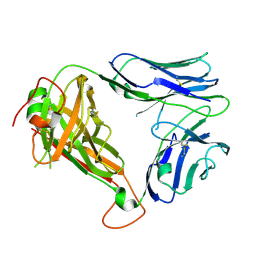 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8 and Beta Chain-TRBV7-8 | | Descriptor: | TCR Alpha Chain-TRAV21-TRAJ8, TCR Beta Chain-TRBV7-8 | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Lim, R.T, Besra, G, Howell, A.R, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
8D4R
 
 | |
5KIH
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA/RNA (5'-D(*TP*TP*AP*G)-R(P*G)-D(P*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
6D2J
 
 | | Beta Carbonic anhydrase in complex with thiocyanate | | Descriptor: | Carbonic anhydrase, POTASSIUM ION, THIOCYANATE ION, ... | | Authors: | Murray, A, Aggarwal, M, Pinard, M, McKenna, R. | | Deposit date: | 2018-04-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Mapping of Anion Inhibitors to beta-Carbonic Anhydrase psCA3 from Pseudomonas aeruginosa.
ChemMedChem, 13, 2018
|
|
5USD
 
 | |
8BIK
 
 | | Crystal structure of human AMPK heterotrimer in complex with allosteric activator C455 | | Descriptor: | (3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-[[6-chloranyl-5-[4-[4-[[dimethyl(oxidanyl)-$l^{4}-sulfanyl]amino]phenyl]phenyl]-3~{H}-imidazo[4,5-b]pyridin-2-yl]oxy]-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]furan-3-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Schimpl, M, Mather, K.M, Boland, M.L, Rivers, E.L, Srivastava, A, Hemsley, P, Robinson, J, Wan, P.T, Hansen, J, Read, J.A, Trevaskis, J.L, Smith, D.M. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direct beta 1/ beta 2 AMPK activation reduces liver steatosis but not fibrosis in a mouse model of non-alcoholic steatohepatitis
Biorxiv, 2024
|
|
5KIB
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA/RNA (5'-D(*TP*TP*AP*G)-R(P*G)-D(P*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5DQQ
 
 | | Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1, ... | | Authors: | Kintzer, A.F, Stroud, R.M. | | Deposit date: | 2015-09-15 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Structure, inhibition and regulation of two-pore channel TPC1 from Arabidopsis thaliana
Nature, 531, 2016
|
|
8BRH
 
 | | Co-crystal structure of She4 with Myo4 peptide | | Descriptor: | KLLA0E16699p, Myo4 peptide (LYS-PHE-ILE-VAL-SER-HIS-TYR) | | Authors: | Arnese, R, Gudino, R, Meinhart, A, Clausen, T. | | Deposit date: | 2022-11-23 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | UNC-45 assisted myosin folding depends on a conserved FX 3 HY motif implicated in Freeman Sheldon Syndrome.
Nat Commun, 15, 2024
|
|
6FZA
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
1LN6
 
 | | STRUCTURE OF BOVINE RHODOPSIN (Metarhodopsin II) | | Descriptor: | RETINAL, RHODOPSIN | | Authors: | Choi, G, Landin, J, Galan, J.F, Birge, R.R, Albert, A.D, Yeagle, P.L. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin.
Biochemistry, 41, 2002
|
|
4WY9
 
 | |
