4MJP
 
 | | E. coli sliding clamp in complex with (R)-Vedaprofen | | Descriptor: | (2R)-2-(4-cyclohexylnaphthalen-1-yl)propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
6HI6
 
 | | The ATAD2 bromodomain in complex with compound 9 | | Descriptor: | (2~{R})-2-azanyl-~{N}-(4-ethanoyl-5-pyridin-3-yl-1,3-thiazol-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
8DEG
 
 | | Crystal structure of DLK in complex with inhibitor DN0011197 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 12, methyl (1S,4S)-5-{(4P)-4-[5-amino-6-(difluoromethoxy)pyrazin-2-yl]-6-[(1R,4R)-2-azabicyclo[2.1.1]hexan-2-yl]pyridin-2-yl}-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Srivastava, A, Lexa, K, de Vicente, J. | | Deposit date: | 2022-06-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of Potent and Selective Dual Leucine Zipper Kinase/Leucine Zipper-Bearing Kinase Inhibitors with Neuroprotective Properties in In Vitro and In Vivo Models of Amyotrophic Lateral Sclerosis.
J.Med.Chem., 65, 2022
|
|
3VPH
 
 | | L-lactate dehydrogenase from Thermus caldophilus GK24 complexed with oxamate, NADH and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Arai, K, Ohno, T, Miyanaga, A, Fushinobu, S, Taguchi, H. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The core of allosteric motion in Thermus caldophilus L-lactate dehydrogenase.
J.Biol.Chem., 2014
|
|
3RO1
 
 | |
7LKX
 
 | | 1.60 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3e | | Descriptor: | (1R,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
6RGY
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
7LKU
 
 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3b (deuterated analog of inhibitor 2a) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKS
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2f | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
2ZYA
 
 | | Dimeric 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, decarboxylating | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
5J05
 
 | |
7LKV
 
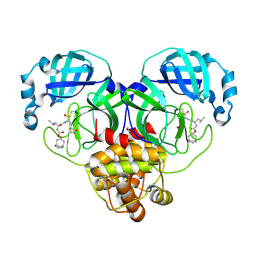 | | 1.55 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
2WLX
 
 | |
6BE2
 
 | |
7LKT
 
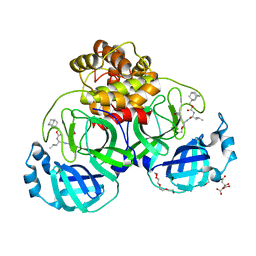 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | Descriptor: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
4XH1
 
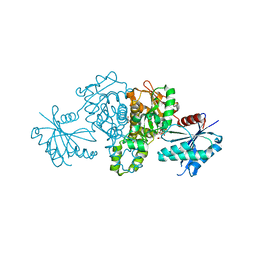 | | Crystal structure of Salmonella typhimurium propionate kinase in complex with AMPPNP and propionate | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROPANOIC ACID, ... | | Authors: | Murthy, A.M.V, Mathivanan, S, Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2015-01-04 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of substrate- and nucleotide-bound propionate kinase from Salmonella typhimurium: substrate specificity and phosphate-transfer mechanism
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7LKR
 
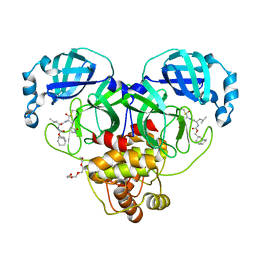 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2a | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
6RJ6
 
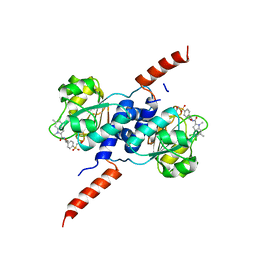 | | Crystal structure of PHGDH in complex with BI-4924 | | Descriptor: | 2-[4-[(1~{S})-1-[[4,5-bis(chloranyl)-1,6-dimethyl-indol-2-yl]carbonylamino]-2-oxidanyl-ethyl]phenyl]sulfonylethanoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
7LKW
 
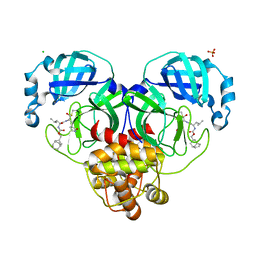 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3d (deuterated analog of inhibitor 3c) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
8CZH
 
 | | Human BAK in complex with the dM2 peptide | | Descriptor: | Bcl-2 homologous antagonist/killer, DM2 peptide | | Authors: | Aguilar, F, Keating, A.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization.
Structure, 31, 2023
|
|
7TLI
 
 | | THERMOLYSIN (90% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-30 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
4D69
 
 | | SOYBEAN AGGLUTININ FROM GLYCINE MAX IN COMPLEX WITH THE ANTIGEN Tn | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LECTIN, ... | | Authors: | Madariaga, D, Martinez-Saez, N, Somovilla, V.J, Coelho, H, Valero-Gonzalez, J, Castro-Lopez, J, Asensio, J.L, Jimenez-Barbero, J, Busto, J.H, Avenoza, A, Marcelo, F, Hurtado-Guerrero, R, Corzana, F, Peregrina, J.M. | | Deposit date: | 2014-11-10 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Detection of Tumor-Associated Glycopeptides by Lectins: The Peptide Context Modulates Carbohydrate Recognition.
Acs Chem.Biol., 10, 2015
|
|
2ZZU
 
 | | Human Factor VIIA-Tissue Factor Complexed with ethylsulfonamide-D-5-(3-carboxybenzyloxy)-Trp-Gln-p-aminobenzamidine | | Descriptor: | 3-[[3-[(2R)-3-[[(2S)-5-amino-1-[(4-carbamimidoylphenyl)methylamino]-1,5-dioxo-pentan-2-yl]amino]-2-(ethylsulfonylamino)-3-oxo-propyl]-1H-indol-5-yl]oxymethyl]benzoic acid, CALCIUM ION, Factor VII heavy chain, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Sato, H, Ohta, M, Kozono, T. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of peptidomimetic factor VIIa inhibitors
Chem.Pharm.Bull., 58, 2010
|
|
4HNF
 
 | | Crystal structure of ck1d in complex with pf4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform delta | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
6RK4
 
 | | Lysostaphin SH3b P4-G5 complex, synchrotron dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, 1,2-ETHANEDIOL, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
