5CUA
 
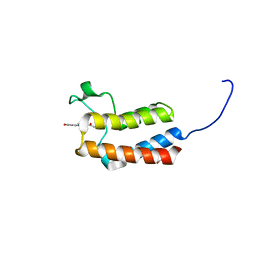 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 1-Acetyl-4-(4-hydroxyphenyl)piperazine (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(4-hydroxyphenyl)piperazin-1-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 1-Acetyl-4-(4-hydroxyphenyl)piperazine (SGC - Diamond I04-1 fragment screening)
To be published
|
|
3ZIT
 
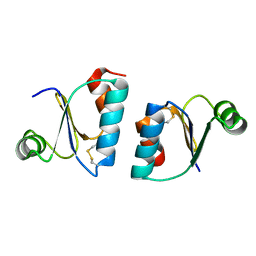 | |
1SZP
 
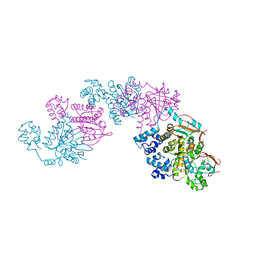 | | A Crystal Structure of the Rad51 Filament | | Descriptor: | DNA repair protein RAD51, SULFATE ION | | Authors: | Conway, A.B, Lynch, T.W, Zhang, Y, Fortin, G.S, Symington, L.S, Rice, P.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of a Rad51 filament.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4RG2
 
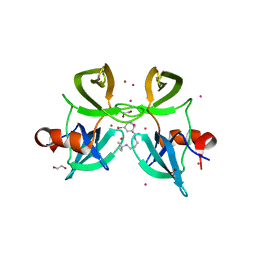 | | Tudor Domain of Tumor suppressor p53BP1 with small molecule ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(tert-butylamino)propyl]benzamide, Tumor suppressor p53-binding protein 1, ... | | Authors: | Dong, A, Mader, P, James, L, Perfetti, M, Tempel, W, Frye, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1.
ACS Chem. Biol., 10, 2015
|
|
1T2F
 
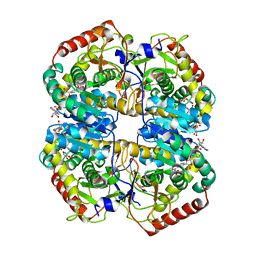 | | Human B lactate dehydrogenase complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | | Descriptor: | 4-HYDROXY-1,2,5-OXADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase B chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
2J1A
 
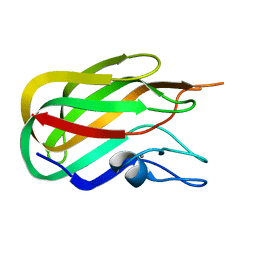 | |
1T2D
 
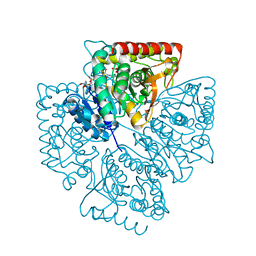 | | Plasmodium falciparum lactate dehydrogenase complexed with NAD+ and oxalate | | Descriptor: | GLYCEROL, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
7B2G
 
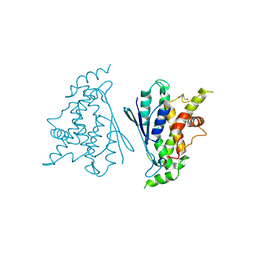 | | Crystal structure of R120Q GDAP1 mutant | | Descriptor: | GLYCEROL, Ganglioside-induced differentiation-associated protein 1 | | Authors: | Nguyen, G.T.T, Sutinen, A, Kursula, P. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conserved intramolecular networks in GDAP1 are closely connected to CMT-linked mutations and protein stability.
Plos One, 18, 2023
|
|
3EG0
 
 | | Crystal structure of the N114T mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
6W8Z
 
 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | Nonstructural polyprotein, methyl 2-oxo-1,2-dihydroquinazoline-4-carboxylate | | Authors: | Zhang, S, Garzan, A, Pathak, A.K, Augelli-Szafran, C.E, Wu, M. | | Deposit date: | 2020-03-21 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
1TGL
 
 | | A SERINE PROTEASE TRIAD FORMS THE CATALYTIC CENTRE OF A TRIACYLGLYCEROL LIPASE | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1990-02-05 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A serine protease triad forms the catalytic centre of a triacylglycerol lipase.
Nature, 343, 1990
|
|
5V28
 
 | | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Dornevil, K, Liu, F, Liu, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To Be Published
|
|
3ZTE
 
 | |
3EKY
 
 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
7TCR
 
 | | Methanobactin biosynthetic protein complex of MbnB and MbnC from Methylosinus trichosporium OB3b at 2.62 Angstrom resolution | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Park, Y, Reyes, R.M, Rosenzweig, A.C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3EMP
 
 | |
1JJ4
 
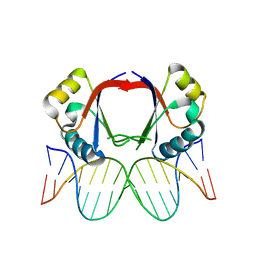 | | Human papillomavirus type 18 E2 DNA-binding domain bound to its DNA target | | Descriptor: | E2 binding site DNA, Regulatory protein E2 | | Authors: | Kim, S.-S, Tam, J.K, Wang, A.F, Hegde, R.S. | | Deposit date: | 2001-07-03 | | Release date: | 2001-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of DNA target discrimination by papillomavirus E2 proteins.
J.Biol.Chem., 275, 2000
|
|
2IG8
 
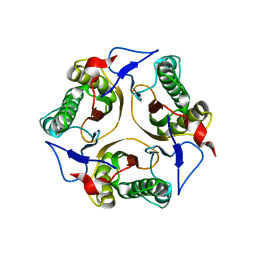 | | Crystal structure of a Protein of Unknown Function PA3499 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical protein PA3499 | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a hypothetical protein PA3499 from Pseudomonas aeruginosa
To be Published
|
|
3EHS
 
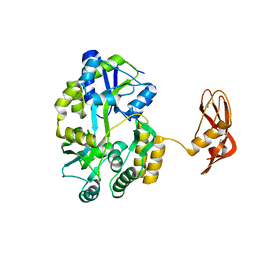 | |
1JJH
 
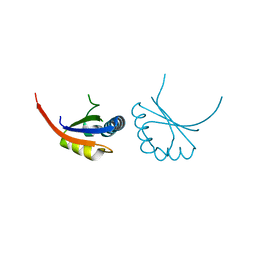 | |
7B3R
 
 | |
4IC7
 
 | | Crystal structure of the ERK5 kinase domain in complex with an MKK5 binding fragment | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 5, Mitogen-activated protein kinase 7, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gogl, G, Remenyi, A. | | Deposit date: | 2012-12-10 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism for the specific assembly and activation of the extracellular signal regulated kinase 5 (ERK5) module.
J.Biol.Chem., 288, 2013
|
|
6W91
 
 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | Nonstructural polyprotein, methyl 2-oxo-2,5-dihydropyrimidine-4-carboxylate | | Authors: | Zhang, S, Garzan, A, Pathak, A.K, Augelli-Szafran, C.E, Wu, M. | | Deposit date: | 2020-03-21 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
1TIP
 
 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, PHOSPHOENZYME INTERMEDIATE OF FRU-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.-H, Olson, T.W, Ogata, C.M, Levitt, D.G, Banaszak, L.J, Lange, A.J. | | Deposit date: | 1997-05-28 | | Release date: | 1998-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
Nat.Struct.Biol., 4, 1997
|
|
2IHK
 
 | |
