8SVU
 
 | | Crystal structure of the L428V mutant of pregnane X receptor ligand binding domain in apo form | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1 | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
5J18
 
 | | Solution structure of Ras Binding Domain (RBD) of B-Raf complexed with Rigosertib (Complex I) | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
5FZP
 
 | | Structure of the dispase autolysis inducing protein from Streptomyces mobaraensis | | Descriptor: | CALCIUM ION, DISPASE AUTOLYSIS-INDUCING PROTEIN, GLYCEROL | | Authors: | Schmelz, S, Fiebig, D, Beck, J, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2016-03-15 | | Release date: | 2016-08-10 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Dispase Autolysis Inducing Protein from Streptomyces Mobaraensis and Glutamine Cross-Linking Sites for Transglutaminase
J.Biol.Chem., 291, 2016
|
|
6HEV
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative AT061 | | Descriptor: | 4-methyl-3-[(2-pyridin-3-ylpyrido[2,3-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
6DLH
 
 | | Endo-fucoidan hydrolase MfFcnA4 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION, ... | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
8Q1T
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with IOTA739 | | Descriptor: | 1,10-PHENANTHROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Lund, B.A, Boronat, P, Hennig, S, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
5FXZ
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with Citrate
To be Published
|
|
5G13
 
 | |
6RAW
 
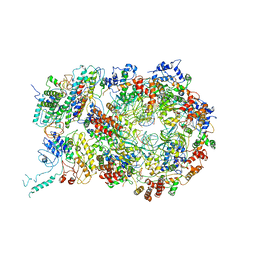 | | D. melanogaster CMG-DNA, State 1A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2019-09-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
7JH3
 
 | | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP | | Descriptor: | 4-aminobutyrate aminotransferase PuuE, DI(HYDROXYETHYL)ETHER | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP
To Be Published
|
|
6X67
 
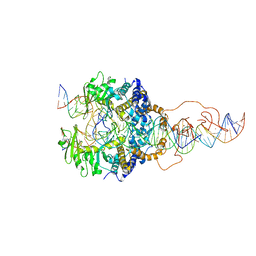 | | Cryo-EM structure of piggyBac transposase strand transfer complex (STC) | | Descriptor: | CALCIUM ION, DNA (37-MER), DNA (47-MER), ... | | Authors: | Chen, Q, Hickman, A.B, Dyda, F. | | Deposit date: | 2020-05-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
5J5I
 
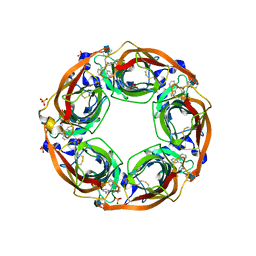 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with 4-(2-amino-6-{bis[(pyridin-2-yl)methyl]amino}pyrimidin-4-yl)phenol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-amino-6-{bis[(pyridin-2-yl)methyl]amino}pyrimidin-4-yl)phenol, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, G.A, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
8T5Z
 
 | |
8TOK
 
 | |
4ERJ
 
 | |
5B6V
 
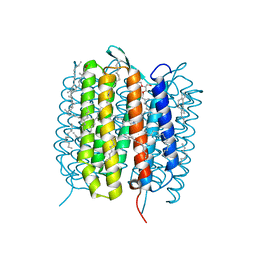 | | A three dimensional movie of structural changes in bacteriorhodopsin: resting state structure | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nango, E, Royant, A, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
7JIS
 
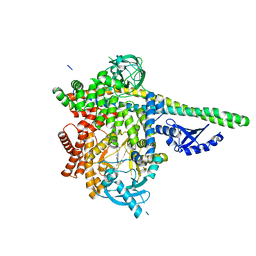 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6HDF
 
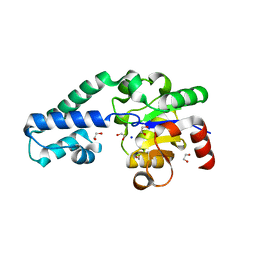 | | D170N variant of beta-phosphoglucomutase from Lactococcus lactis in an open conformer to 1.4 A. | | Descriptor: | 1,2-ETHANEDIOL, Beta-phosphoglucomutase, SODIUM ION | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6R7I
 
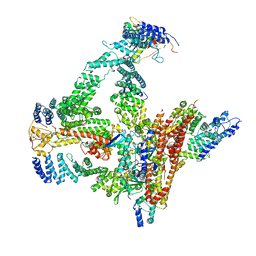 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
8T6Z
 
 | |
6HDP
 
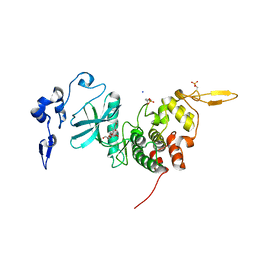 | | Human DYRK2 bound to Scorzodihydrostilbene A | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, SODIUM ION, Scorzodihydrostilbene A | | Authors: | Elkins, J.M, Soundararajan, M, Proksch, P, Meijer, L, Vollmar, M, Krojer, T, Bountra, C, Edwards, A.M, Arrowsmith, C, Knapp, S. | | Deposit date: | 2018-08-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DYRK2 bound to Scorzodihydrostilbene A
To Be Published
|
|
5FXW
 
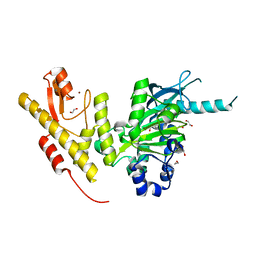 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with fumarate | | Descriptor: | 1,2-ETHANEDIOL, FUMARIC ACID, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with Fumarate
To be Published
|
|
7JJP
 
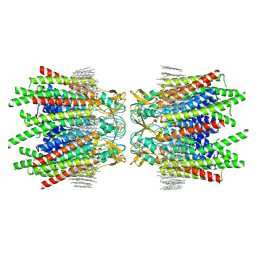 | | Sheep Connexin-50 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
6UI7
 
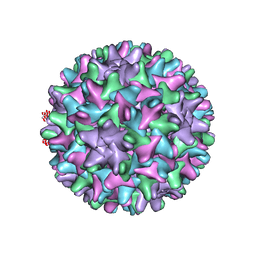 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6R5X
 
 | |
