5HYT
 
 | |
184L
 
 | |
6VP7
 
 | |
6RM2
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ATP, IMP, Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, INOSINIC ACID, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, IMP, Magnesium at 2.5 Angstrom resolution
To Be Published
|
|
5Z3E
 
 | | Glycosidase E335A | | Descriptor: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
6RMK
 
 | | Bacteriorhodopsin, dark state, cell 2, refined using the same protocol as sub-ps time delays | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6YQJ
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with open-chain Fasudil-derivative 2-[isoquinolin-5-ylsulfonyl(propyl)amino]ethylazanium (soaked) | | Descriptor: | 2-[isoquinolin-5-ylsulfonyl(propyl)amino]ethylazanium, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
4TZD
 
 | | Crystal structure of Canavalia maritima lectin (ConM) complexed with interleukin - 1 beta primer | | Descriptor: | Concanavalin-A, DNA (5'-D(P*CP*G)-3'), DNA (5'-D(P*TP*C)-3') | | Authors: | Vieira, D.B.H.A, Delatorre, P, Rocha, B.A.M, Teixeira, C.S, Silva-Filho, J.C, Lima, E.M, Nobrega, R.B, Cavada, B.S. | | Deposit date: | 2014-07-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Canavalia maritima lectin (ConM) complexed with interleukin 1 - beta primer
To Be Published
|
|
6W0A
 
 | | Open-gate KcsA soaked in 1 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.237 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
7BTY
 
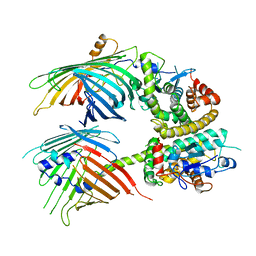 | | The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
5KSW
 
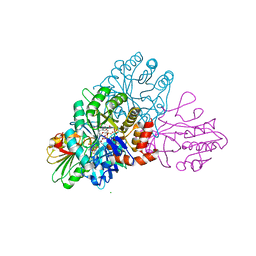 | | DHODB-I74D mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2016-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Isoleucine 74 Plays a Key Role in Controlling Electron Transfer Between Lactococcus lactis Dihydroorotate Dehydrogenase 1B Subunits
To Be Published
|
|
6RUG
 
 | |
6W0I
 
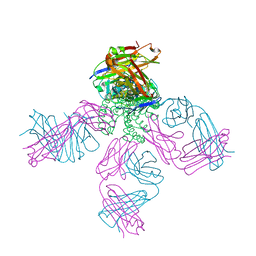 | | Closed-gate KcsA soaked in 10mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6RUN
 
 | |
6W0Q
 
 | | APE1 endonuclease product complex D148E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-02 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
1A3L
 
 | |
4ZOP
 
 | | Co-crystal Structure of Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitor | | Descriptor: | (2S,3R)-N~1~-(8-tert-butyl-4,5-dihydro[1,3]thiazolo[4,5-h]quinazolin-2-yl)-3-methylpyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Co-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitorCo-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitor
To Be Published
|
|
8OPR
 
 | |
8CXO
 
 | | Cryo-EM structure of the unliganded mSMO-PGS2 in a lipidic environment | | Descriptor: | CHOLESTEROL, Smoothened homolog, GlgA glycogen synthase chimera | | Authors: | Zhang, K, Wu, H, Hoppe, N, Manglik, A, Cheng, Y. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
7TYA
 
 | | Cryo-EM structure of human Anion Exchanger 1 modified with Diethyl Pyrocarbonate (DEPC) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5FLR
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
6C6P
 
 | | Human squalene epoxidase (SQLE, squalene monooxygenase) structure with FAD and NB-598 | | Descriptor: | (2E)-N-({3-[([3,3'-bithiophen]-5-yl)methoxy]phenyl}methyl)-N-ethyl-6,6-dimethylhept-2-en-4-yn-1-amine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Padyana, A.K, Jin, L. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase.
Nat Commun, 10, 2019
|
|
7BQU
 
 | | Cereblon in complex with SALL4 and (S)-thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, Sal-like protein 4, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
7TY8
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Niflumic Acid | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6W43
 
 | | APE1 AP-endonuclease product complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
