4RG9
 
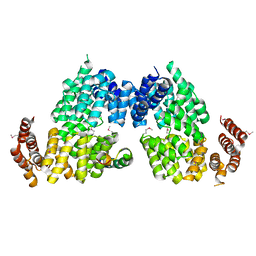 | | Crystal structure of APC3-APC16 complex (Selenomethionine Derivative) | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
6CGS
 
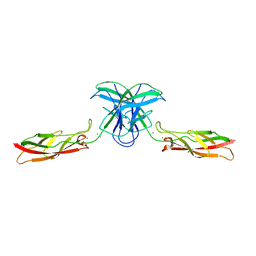 | | mouse cadherin-7 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-7, GLYCEROL | | Authors: | Brasch, J, Harrison, O.J, Kaczynska, A, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6SSK
 
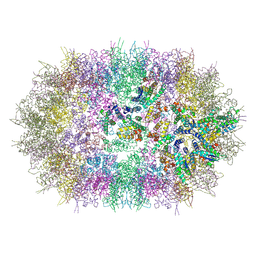 | |
2WXN
 
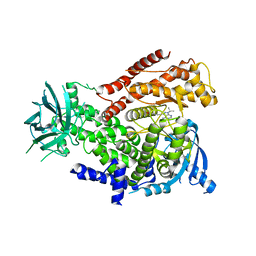 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with DL07. | | Descriptor: | 3-{[4-amino-1-(1-methylethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]ethynyl}phenol, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
6NIG
 
 | | Crystal structure of the human TLR2-Diprovocim complex | | Descriptor: | (3S,4S,3'S,4'S)-1,1'-(1,4-phenylenedicarbonyl)bis{N~3~,N~4~-bis[(1S,2R)-2-phenylcyclopropyl]pyrrolidine-3,4-dicarboxami de}, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Beutler, B.A, Tomchick, D.R, Su, L. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim.
J. Med. Chem., 62, 2019
|
|
5HKY
 
 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
9CZ1
 
 | |
5TK0
 
 | | Lysozyme structure collected with 3D printed polymer mounts | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kilah, N.L, MacDonald, N, Bunton, G, Park, A.Y, Breadmore, M.C. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | 3D Printed Micrometer-Scale Polymer Mounts for Single Crystal Analysis.
Anal. Chem., 89, 2017
|
|
6VVO
 
 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
7JJM
 
 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS | | Descriptor: | CHLORIDE ION, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McGuaig, R, Tan, H.Y.A, Hardy, C, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
8CL4
 
 | |
5FHC
 
 | |
8CL2
 
 | |
4Z4K
 
 | |
4Z4W
 
 | | Crystal structure of GII.10 P domain in complex with 4.7mM fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, beta-L-fucopyranose | | Authors: | Koromyslova, A.D, Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
2WXM
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with DL06. | | Descriptor: | 1-(1-METHYLETHYL)-3-(PYRIDIN-3-YLETHYNYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-AMINE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
5N36
 
 | |
4Z6I
 
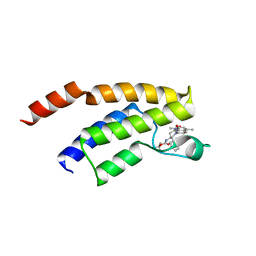 | | Crystal structure of BRD9 bromodomain in complex with a substituted valerolactam quinolone ligand | | Descriptor: | Bromodomain-containing protein 9, tert-butyl [(2R,3S)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxo-2-phenylpiperidin-3-yl]carbamate | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6CEI
 
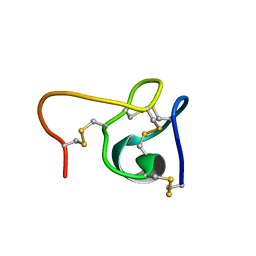 | |
5BMQ
 
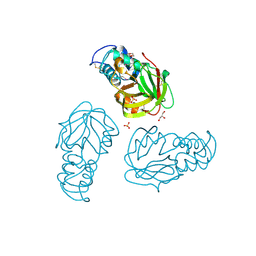 | | Crystal structure of L,D-transpeptidase (Yku) from Stackebrandtia nassauensis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ErfK/YbiS/YcfS/YnhG family protein, GLYCEROL, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of L,D-transpeptidases (Yku)
To be Published
|
|
6CL8
 
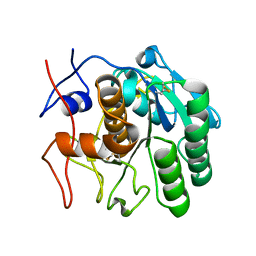 | | 2.00 A MicroED structure of proteinase K at 2.6 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
2WXH
 
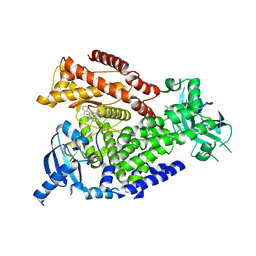 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with SW14. | | Descriptor: | 2-{[4-amino-3-(3-fluoro-4-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
6FPT
 
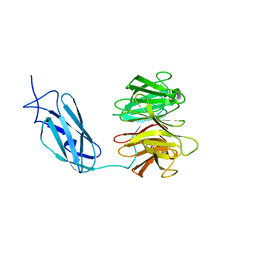 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains | | Descriptor: | E3 ubiquitin-protein ligase TRIM71 | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
8CL0
 
 | |
6HM3
 
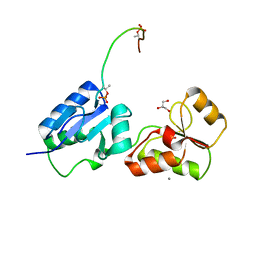 | | Crystal structure of Rad4 BRCT1,2 in complex with a Sld3 phosphopeptide | | Descriptor: | CALCIUM ION, DNA replication regulator sld3, GLYCEROL, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77263618 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
