4ZZW
 
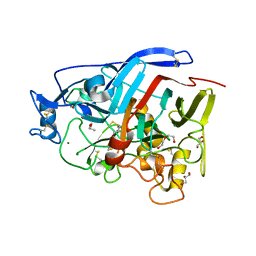 | | Geotrichum candidum Cel7A structure complex with cellobiose at 1.5A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE I, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
5A3C
 
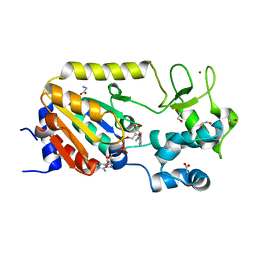 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
4UG2
 
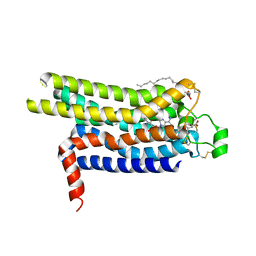 | | Thermostabilised HUMAN A2a Receptor with CGS21680 bound | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[P-(2-CARBOXYETHYL)PHENYLETHYL-AMINO]-5'-N-ETHYLCARBOXAMIDO ADENOSINE, THERMOSTABILISED HUMAN A2A RECEPTOR | | Authors: | Lebon, G, Edwards, P.C, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-03-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Determinants of Cgs21680 Binding to the Human Adenosine A2A Receptor.
Mol.Pharmacol., 87, 2015
|
|
5A3Y
 
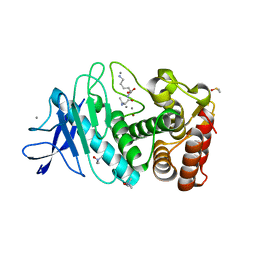 | | SAD structure of Thermolysin obtained by multi crystal data collection | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-04 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6FYZ
 
 | | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor | | Descriptor: | (2~{S})-2-(2-fluorophenyl)-2-[4-(2-methylpyrimidin-5-yl)phenyl]-~{N}-oxidanyl-ethanamide, Histone deacetylase 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Maillard, M.C, Dominguez, C. | | Deposit date: | 2018-03-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5FF0
 
 | | HydE from T. maritima in complex with S-adenosyl-L-cysteine and methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, Fe4-Se4 cluster, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
1AMD
 
 | |
6SXT
 
 | | GH54 a-l-arabinofuranosidase soaked with aziridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
6WOP
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii | | Descriptor: | 4-aminobutyrate transaminase, CHLORIDE ION, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii
To Be Published
|
|
6C0L
 
 | | Crystal structure of HIV-1 E138K mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
5ACN
 
 | |
6G3F
 
 | | Crystal structure of EDDS lyase in complex with fumarate | | Descriptor: | Argininosuccinate lyase, DI(HYDROXYETHYL)ETHER, FUMARIC ACID | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
5FIV
 
 | | STRUCTURAL STUDIES OF HIV AND FIV PROTEASES COMPLEXED WITH AN EFFICIENT INHIBITOR OF FIV PR | | Descriptor: | RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Li, M, Lee, T, Morris, G, Laco, G, Wong, C, Olson, A, Elder, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-12-02 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
4ZXI
 
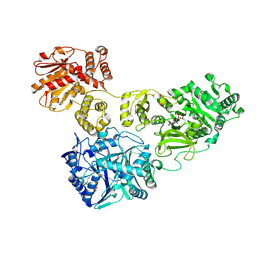 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase bound to AMP and Glycine | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
5A8G
 
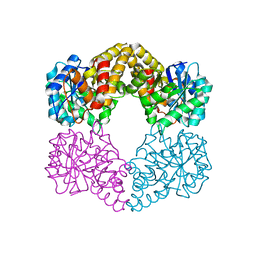 | | Crystal structure of the wild-type Staphylococcus aureus N- acetylneurminic acid lyase in complex with fluoropyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Stockwell, J, Daniels, A.D, Windle, C.L, Harman, T, Woodhall, T, Trinh, C.H, Lebel, T, Pearson, A.R, Mulholland, K, Berry, A, Nelson, A. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evaluation of Fluoropyruvate as Nucleophile in Reactions Catalysed by N-Acetyl Neuraminic Acid Lyase Variants: Scope, Limitations and Stereoselectivity.
Org.Biomol.Chem., 14, 2016
|
|
6GA2
 
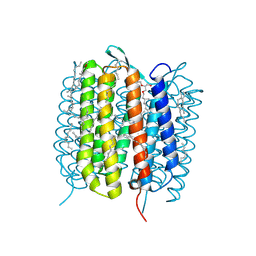 | | Bacteriorhodopsin, dark state, cell 2 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6SZX
 
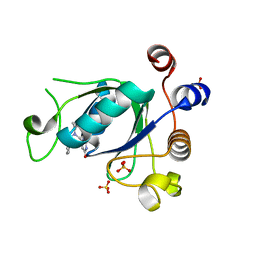 | | Crystal structure of YTHDC1 with fragment 11 (DHU_DC1_128) | | Descriptor: | 6-[[cyclopropyl-(phenylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
5JAM
 
 | | Yersinia pestis FabV variant T276V | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
5FNH
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 5-[(3-chloranylphenoxy)methyl]-1,2,4-triaza-3-azanidacyclopenta-1,4-diene, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-11-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
6T0C
 
 | | Crystal structure of YTHDC1 with fragment 26 (DHU_DC1_198) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-2~{H}-indazole-3-carboxamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
5JBT
 
 | | Mesotrypsin in complex with cleaved amyloid precursor like protein 2 inhibitor (APLP2) | | Descriptor: | Amyloid-like protein 2, CALCIUM ION, PRSS3 protein, ... | | Authors: | Kayode, O, Wang, R, Pendlebury, D, Soares, A, Radisky, E.S. | | Deposit date: | 2016-04-13 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Acrobatic Substrate Metamorphosis Reveals a Requirement for Substrate Conformational Dynamics in Trypsin Proteolysis.
J. Biol. Chem., 291, 2016
|
|
1A4L
 
 | | ADA STRUCTURE COMPLEXED WITH DEOXYCOFORMYCIN AT PH 7.0 | | Descriptor: | 2'-DEOXYCOFORMYCIN, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wang, Z, Quiocho, F.A. | | Deposit date: | 1998-01-31 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
Biochemistry, 37, 1998
|
|
6T0M
 
 | | Cationic Trypsin in Complex with a D-Phe-Pro-diaminopyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[[2,6-bis(azanyl)pyridin-4-yl]methyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
7KXL
 
 | | BTK1 SOAKED WITH COMPOUND 5, Y551 IS SEQUESTERED | | Descriptor: | 3-tert-butyl-N-({2-fluoro-4-[2-(1-methyl-1H-pyrazol-4-yl)-1H-imidazo[4,5-b]pyridin-7-yl]phenyl}methyl)-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
