6EC0
 
 | | Crystal structure of the wild-type heterocomplex between coil 1B domains of human intermediate filament proteins keratin 1 (KRT1) and keratin 10 (KRT10) | | Descriptor: | CADMIUM ION, COBALT (II) ION, Keratin 1, ... | | Authors: | Eldirany, S.A, Lomakin, I.B, Bunick, C.G. | | Deposit date: | 2018-08-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly.
Embo J., 38, 2019
|
|
7SAH
 
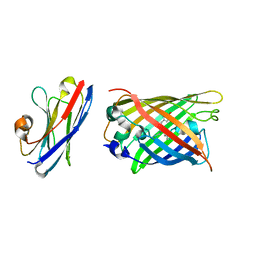 | |
6HYF
 
 | |
4ZAR
 
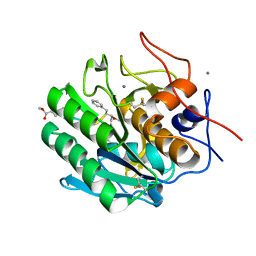 | | Crystal Structure of Proteinase K from Engyodontium albuminhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE, bound form, ... | | Authors: | Sawaya, M.R, Cascio, D, Collazo, M, Bond, C, Cohen, A, DeNicola, A, Eden, K, Jain, K, Leung, C, Lubock, N, McCormick, J, Rosinski, J, Spiegelman, L, Athar, Y, Tibrewal, N, Winter, J, Solomon, S. | | Deposit date: | 2015-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Proteinase K from Engyodontium album inhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution
to be published
|
|
6RAH
 
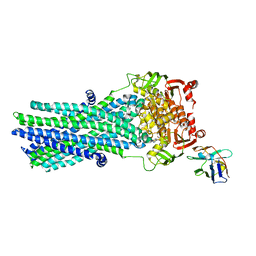 | | Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding and permease protein, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
5YZO
 
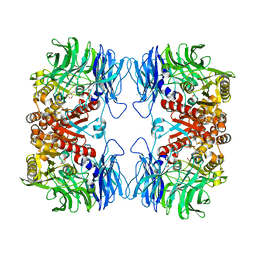 | | Crystal structure of S9 peptidase mutant (S514A) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, DIMETHYL SULFOXIDE, ... | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
4UB3
 
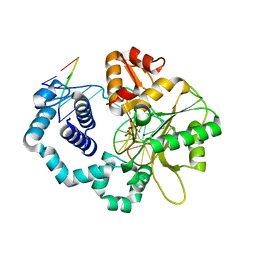 | | DNA polymerase beta closed product complex with a templating cytosine and 8-oxodGMP, 60 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
6RAS
 
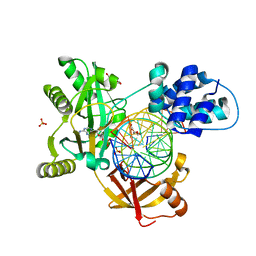 | | Pmar-Lig_Pre. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA, ... | | Authors: | Leiros, H.K.S, Williamson, A. | | Deposit date: | 2019-04-07 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
8HCJ
 
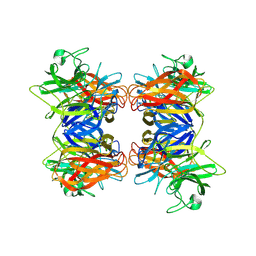 | | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans | | Descriptor: | CALCIUM ION, Xylan 1,4-beta-xylosidase | | Authors: | Vishwakarma, P, Sachdeva, E, Goyal, A, Ethayathulla, A.S, Das, U, Kaur, P. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans
To Be Published
|
|
6R2G
 
 | | Crystal structure of a single-chain protein mimetic of the gp41 NHR trimer in complex with the synthetic CHR peptide C34 | | Descriptor: | Envelope glycoprotein gp160, PHOSPHATE ION, Single-chain protein mimetics of the N-terminal heptad-repeat region of gp41 | | Authors: | Camara-Artigas, A, Conejero-Lara, F, Jurado, S, Cano-Munoz, M, Morel, B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Thermodynamic Analysis of HIV-1 Fusion Inhibition Using Small gp41 Mimetic Proteins.
J.Mol.Biol., 431, 2019
|
|
6W8Z
 
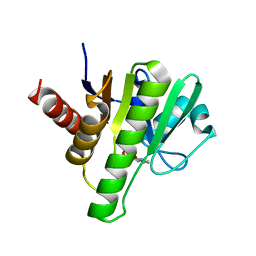 | | Co-crystal structures of CHIKV nsP3 macrodomain with pyrimidone fragments | | Descriptor: | Nonstructural polyprotein, methyl 2-oxo-1,2-dihydroquinazoline-4-carboxylate | | Authors: | Zhang, S, Garzan, A, Pathak, A.K, Augelli-Szafran, C.E, Wu, M. | | Deposit date: | 2020-03-21 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design.
Plos One, 16, 2021
|
|
6JPL
 
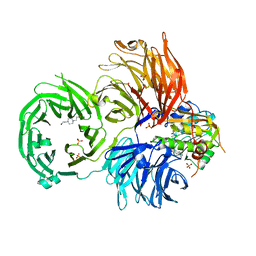 | | The X-ray structure of yeast tRNA methyltransferase Trm7-Trm734 in complex with S-adenosyl-L-methionine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Shimizu, N, Hori, H. | | Deposit date: | 2019-03-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
5IUS
 
 | | Crystal structure of human PD-L1 in complex with high affinity PD-1 mutant | | Descriptor: | CHLORIDE ION, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Pascolutti, R, Sun, X, Kao, J, Maute, R, Ring, A.M, Bowman, G.R, Kruse, A.C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Structure and Dynamics of PD-L1 and an Ultra-High-Affinity PD-1 Receptor Mutant.
Structure, 24, 2016
|
|
5APO
 
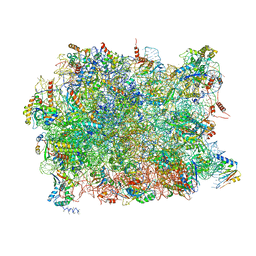 | | Structure of the yeast 60S ribosomal subunit in complex with Arx1, Alb1 and C-terminally tagged Rei1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Greber, B.J, Gerhardy, S, Leitner, A, Leibundgut, M, Salem, M, Boehringer, D, Leulliot, N, Aebersold, R, Panse, V.G, Ban, N. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation.
Cell(Cambridge,Mass.), 164, 2016
|
|
4ZCH
 
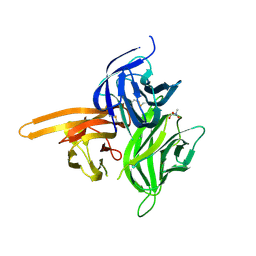 | | Single-chain human APRIL-BAFF-BAFF Heterotrimer | | Descriptor: | TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, Tumor necrosis factor ligand superfamily member 13,Tumor necrosis factor ligand superfamily member 13B,Tumor necrosis factor ligand superfamily member 13B | | Authors: | Lammens, A, Jiang, X, Maskos, K, Schneider, P. | | Deposit date: | 2015-04-16 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Stoichiometry of Heteromeric BAFF and APRIL Cytokines Dictates Their Receptor Binding and Signaling Properties.
J.Biol.Chem., 290, 2015
|
|
1ADD
 
 | |
6CAC
 
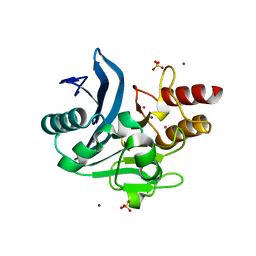 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
5J8H
 
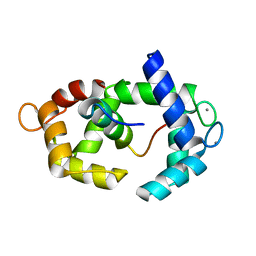 | | Structure of calmodulin in a complex with a peptide derived from a calmodulin-dependent kinase | | Descriptor: | CALCIUM ION, Calmodulin, Eukaryotic elongation factor 2 kinase | | Authors: | Alphonse, S, Lee, K, Piserchio, A, Tavares, C.D.J, Giles, D.H, Wellmann, R.M, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Eukaryotic Elongation Factor 2 Kinase by Calmodulin.
Structure, 24, 2016
|
|
6S59
 
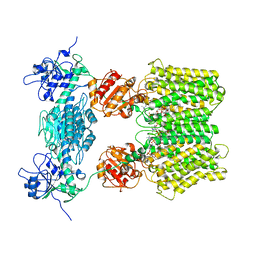 | | Structure of ovine transhydrogenase in the apo state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Nicotinamide nucleotide transhydrogenase | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2019-07-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and mechanism of mitochondrial proton-translocating transhydrogenase.
Nature, 573, 2019
|
|
8TU9
 
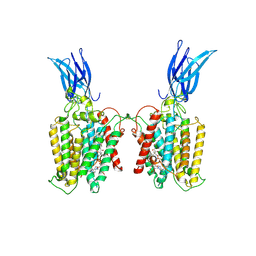 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N -acetyltransferase (HGSNAT).
Elife, 13, 2024
|
|
6S5Z
 
 | | Structure of Rib R28N from Streptococcus pyogenes | | Descriptor: | SODIUM ION, Surface protein R28 | | Authors: | Whelan, F, Griffiths, S.C, Whittingham, J.L, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6W5K
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5g | | Descriptor: | 3C-LIKE PROTEASE, N~2~-{[2-(3-chlorophenyl)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
5Z6O
 
 | | Crystal structure of Penicillium cyclopium protease | | Descriptor: | CALCIUM ION, phenylmethanesulfonic acid, protease | | Authors: | Ko, T.-P, Koszelak, S, Ng, J, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystallographic structure of the subtilisin protease from Penicillium cyclopium.
Biochemistry, 36, 1997
|
|
6GA1
 
 | | Bacteriorhodopsin, dark state, cell 1 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6RK3
 
 | | Solution structure of the ribosome Elongation Factor P (EF-P) from Staphylococcus aureus | | Descriptor: | Elongation factor P | | Authors: | Usachev, K, Fatkhullin, B, Gabdulkhakov, A, Khusainov, I, Golubev, A, Validov, S, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
