4WPK
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form I | | Descriptor: | CITRIC ACID, SODIUM ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6GZ2
 
 | | Crystal Structure of the LeuO Effector Binding Domain | | Descriptor: | HTH-type transcriptional regulator LeuO, MALONATE ION | | Authors: | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | Deposit date: | 2018-07-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
6B0N
 
 | | Crystal structure of the cleavage-independent prefusion HIV Env glycoprotein trimer of the clade A BG505 isolate (NFL construct) in complex with Fabs PGT122 and PGV19 at 3.39 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp140, ... | | Authors: | Sarkar, A, Irimia, A, Wilson, I.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer.
Nat Commun, 9, 2018
|
|
6UJS
 
 | | P-glycoprotein mutant-F728A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
8UM9
 
 | |
7BKY
 
 | | Endothiapepsin structure obtained at 298K with fragment BTB09871 bound from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, PENTAETHYLENE GLYCOL, ... | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endothiapepsin structure obtained at 298K with fragment BTB09871 bound from a dataset collected with JUNGFRAU detector
To Be Published
|
|
4WU7
 
 | | HLA-A24 in complex with HIV-1 Nef134-8(2F) | | Descriptor: | 8-Mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Yamagata, A, Fukai, S, Iwamoto, A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of Nef134-8 epitope presented by HLA-A24
To Be Published
|
|
8P4U
 
 | | Alpha-1-antitrypsin - Sydney variant (G192C) | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Kamuda, K, Lomas, D.A, Irving, J.A. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel pathological mutant reveals the role of torsional flexibility in the serpin breach in adoption of an aggregation-prone intermediate.
Febs J., 291, 2024
|
|
6T92
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6GCZ
 
 | | Laminarin binding SusD-like protein | | Descriptor: | GLYCEROL, Starch-binding associating with outer membrane | | Authors: | Mystowka, A, Robb, C.S, Hehemann, J.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular recognition of the beta-glucans laminarin and pustulan by a SusD-like glycan-binding protein of a marine Bacteroidetes.
FEBS J., 285, 2018
|
|
5N8N
 
 | | Contracted sheath of a Pseudomonas aeruginosa type six secretion system consisting of TssB1 and TssC1 | | Descriptor: | EvpB family type VI secretion protein, Type VI secretion protein, family | | Authors: | Salih, O, He, S, Stach, L, Macdonald, J.T, Planamente, S, Manoli, E, Scheres, S, Filloux, A, Freemont, P.S. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Atomic Structure of Type VI Contractile Sheath from Pseudomonas aeruginosa.
Structure, 26, 2018
|
|
6T8Z
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
7B8W
 
 | | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 1 | | Authors: | Lee, H, Yosaatmadja, Y, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Elkins, J.M. | | Deposit date: | 2020-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470
To Be Published
|
|
5L3Y
 
 | | Designed Artificial Cupredoxins | | Descriptor: | Streptavidin, [CuII(biot-et-dpea)]2+ | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Artificial Cupredoxins.
J.Am.Chem.Soc., 138, 2016
|
|
5L4M
 
 | | Crystal Structure of Human Transthyretin in Complex with 3,5,6-Trichloro-2-pyridinyloxyacetic acid (Triclopyr) | | Descriptor: | SODIUM ION, Transthyretin, Triclopyr | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
8A6T
 
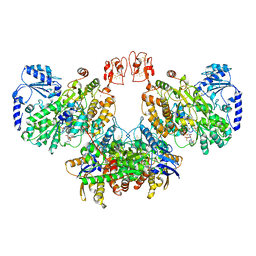 | | Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the reduced state | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, Electron bifurcating hydrogenase subunit HydA1, Electron bifurcating hydrogenase subunit HydB, ... | | Authors: | Kumar, A, Saura, P, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-06-19 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
5N7J
 
 | |
5NBU
 
 | |
8A5E
 
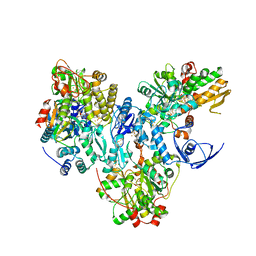 | | Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Acetobacterium woodii in the reduced state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
5KT6
 
 | | Teranry complex of human DNA polymerase iota(1-445) inserting dCMPNPP opposite template G in the presence of Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(P*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
7ZY6
 
 | |
5MDI
 
 | | Crystal structure of TDP-43 N-terminal domain at 2.1 A resolution | | Descriptor: | ACETATE ION, TAR DNA-binding protein 43 | | Authors: | Afroz, T, Hock, E.-M, Ernst, P, Foglieni, C, Jambeau, M, Gilhespy, L, Laferriere, F, Maniecka, Z, Plueckthun, A, Mittl, P, Paganetti, P, Allain, F.H.T, Polymenidou, M. | | Deposit date: | 2016-11-11 | | Release date: | 2017-07-05 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation.
Nat Commun, 8, 2017
|
|
7OT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND inhibitor RMC-282 | | Descriptor: | (R)-N-(1-(6-amino-9H-purin-9-yl)propan-2-yl)-N-(2-phosphonoethyl)glycine, DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
6TL3
 
 | | Crystal structure of an Estrogen Receptor alpha 8-mer phosphopeptide in complex with 14-3-3sigma stabilized by a Pyrrolidone1 derivative | | Descriptor: | 14-3-3 protein sigma, 5-[(2~{S},3~{R})-3-[(~{R})-azanyl(phenyl)methyl]-2-(4-nitrophenyl)-4,5-bis(oxidanylidene)pyrrolidin-1-yl]-2-oxidanyl-benzoic acid, Estrogen receptor | | Authors: | Andrei, S.A, Bosica, F, Ottmann, C, O'Mahony, G. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Design of Drug-Like Protein-Protein Interaction Stabilizers Guided By Chelation-Controlled Bioactive Conformation Stabilization.
Chemistry, 26, 2020
|
|
7OTX
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-257 | | Descriptor: | (S)-2-((3-(6-amino-9H-purin-9-yl)propyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
