6SZ7
 
 | | Crystal structure of YTHDC1 with fragment 5 (DHU_DC1_066) | | Descriptor: | 5,6,7,8-tetrahydro-4~{a}~{H}-quinazoline-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SPM
 
 | | Crystal structure of cAMP-dependent protein kinase A (CHO PKA) in complex with 4-(trifluoromethyl)benzoic acid | | Descriptor: | 4-(trifluoromethyl)benzoic acid, DIMETHYL SULFOXIDE, METHANOL, ... | | Authors: | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2019-09-02 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6VR7
 
 | | Structure of a pseudomurein peptide ligase type C from Methanothermus fervidus | | Descriptor: | ACETOACETIC ACID, GLYCEROL, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry
FEMS Microbes, 2, 2021
|
|
5I6I
 
 | | Crystal structure of a dBCCP-variant of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein,Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.4 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
6VQS
 
 | |
5DH7
 
 | | Two divalent metal ions and conformational changes play roles in the hammerhead ribozyme cleavage reaction-G12A mutant in Mn2+ | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*UP*DC*UP*GP*GP*GP*CP*AP*GP*UP*AP*CP*CP*CP*A)-3', MANGANESE (II) ION, RNA (48-MER) | | Authors: | Mir, A, Chen, J, Neau, D, Golden, B.L. | | Deposit date: | 2015-08-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.064 Å) | | Cite: | Two Divalent Metal Ions and Conformational Changes Play Roles in the Hammerhead Ribozyme Cleavage Reaction.
Biochemistry, 54, 2015
|
|
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
4WT2
 
 | | Co-crystal Structure of MDM2 in Complex with AM-7209 | | Descriptor: | 4-({[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetyl}amino)-2-methoxybenzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of AM-7209, a Potent and Selective 4-Amidobenzoic Acid Inhibitor of the MDM2-p53 Interaction.
J.Med.Chem., 57, 2014
|
|
6SQ1
 
 | | Crystal structure of cAMP-dependent Protein Kinase A (CHO PKA) in complex with Aminofasudil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-diazepan-1-ylsulfonyl)isoquinolin-1-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A crystallographic study of cAMP-dependent Protein Kinase A in complex with different Fasudil-derivatives
To Be Published
|
|
5IB2
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
6YXS
 
 | |
8PNL
 
 | | Outward-open conformation of a Major Facilitator Superfamily (MFS) transporter MHAS2168, a homologue of Rv1410 from M. tuberculosis, in complex with an alpaca nanobody | | Descriptor: | Nb_H2, Putative triacylglyceride transporter | | Authors: | Remm, S, Schoeppe, J, Hutter, C.A.J, Gonda, I, Seeger, M.A. | | Deposit date: | 2023-06-30 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for triacylglyceride extraction from mycobacterial inner membrane by MFS transporter Rv1410.
Nat Commun, 14, 2023
|
|
6CCK
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3-(3-chlorophenyl)-3-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)propanenitrile | | Descriptor: | (3R)-3-(3-chlorophenyl)-3-[(5-methyl-7-oxo-6,7-dihydro[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]propanenitrile, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
5M1G
 
 | |
6SRI
 
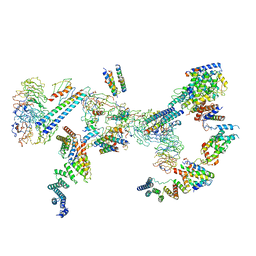 | | Structure of the Fanconi anaemia core complex | | Descriptor: | Fanconi anaemia protein FANCL, Unassigned secondary structure elements (base region, proposed FANCC-FANC-E-FANCF), ... | | Authors: | Shakeel, S, Rajendra, E, Alcon, P, He, S, Scheres, S.H.W, Passmore, L.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the Fanconi anaemia monoubiquitin ligase complex.
Nature, 575, 2019
|
|
8DHE
 
 | |
7ZTV
 
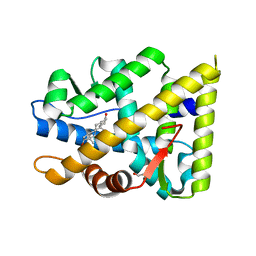 | | Crystal structure of mutant AR-LBD (F755L) bound to dihydrotestosterone | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
6FNQ
 
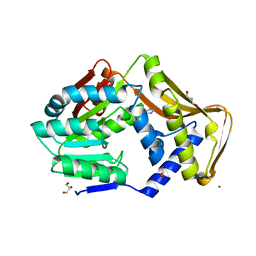 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N,N-trimethylhistidine (hercynine) | | Descriptor: | GLYCEROL, Histidine N-alpha-methyltransferase, MAGNESIUM ION, ... | | Authors: | Vit, A, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition and Regulation of the Ergothioneine Biosynthetic Methyltransferase EgtD.
ACS Chem. Biol., 13, 2018
|
|
7ZTX
 
 | | Crystal structure of mutant AR-LBD (F755V) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZU2
 
 | | Crystal structure of mutant AR-LBD (Q799E) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
5D2I
 
 | |
8A6E
 
 | | 100 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
4WWQ
 
 | | Apo structure of the Grb7 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 7, MALONIC ACID | | Authors: | Watson, G.M, Ambaye, N.D, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
5M3B
 
 | | Structure of cobinamide-bound BtuF mutant W66L, the periplasmic vitamin B12 binding protein in E.coli | | Descriptor: | COB(II)INAMIDE, CYANIDE ION, GLYCEROL, ... | | Authors: | Mireku, S.A, Ruetz, M, Zhou, T, Korkhov, V.M, Kraeutler, B, Locher, K.P. | | Deposit date: | 2016-10-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Change of a Tryptophan Residue in BtuF Facilitates Binding and Transport of Cobinamide by the Vitamin B12 Transporter BtuCD-F.
Sci Rep, 7, 2017
|
|
4WWW
 
 | | Crystal structure of the E. coli ribosome bound to CEM-101 | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S rRNA, 23S rRNA, ... | | Authors: | Dunkle, J.A, Zhang, W, Cate, J.H.D, Mankin, A.S. | | Deposit date: | 2014-11-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis.
Antimicrob. Agents Chemother., 54, 2010
|
|
