9EOY
 
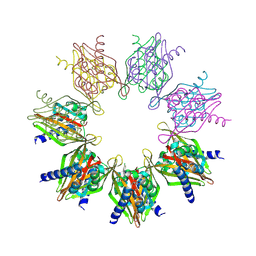 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to PIPA | | Descriptor: | 2-[6-(4-chlorophenyl)imidazo[1,2-b]pyridazin-2-yl]ethanoic acid, ACETATE ION, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Narayanan, D, Larsen, A.S.G, Solbak, S.M.O, Wellendorph, P, Gee, C.L, Kastrup, J.S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced CaMKII alpha hub Trp403 flip, hub domain stacking, and modulation of kinase activity.
Protein Sci., 33, 2024
|
|
9FQ9
 
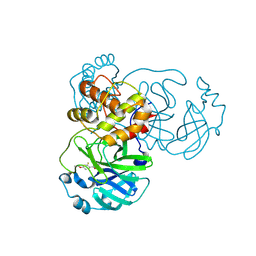 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor PSB-21110 (compound 29b in publication) | | Descriptor: | (5-chloranylpyridin-3-yl) 4-ethoxy-2-fluoranyl-benzoate, BROMIDE ION, Non-structural protein 11 | | Authors: | Strater, N, Claff, T, Sylvester, K, Oneto, A, Guetschow, M, Mueller, C.E. | | Deposit date: | 2024-06-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Nonpeptidic Irreversible Inhibitors of SARS-CoV-2 Main Protease with Potent Antiviral Activity.
J.Med.Chem., 67, 2024
|
|
9BC2
 
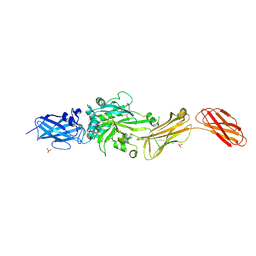 | | Transglutaminase 2 - Open State | | Descriptor: | CHLORIDE ION, HB-225 (gluten peptidomimetic TG2 inhibitor), Protein-glutamine gamma-glutamyltransferase 2, ... | | Authors: | Mathews, I.I, Sewa, A, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BRS
 
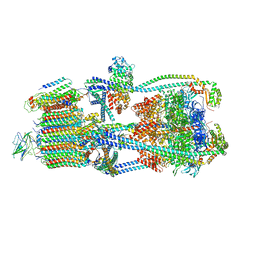 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BBH
 
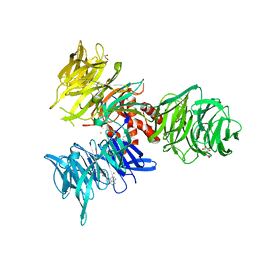 | | Co-crystal structure of human DDB1 bound to fragment UB028670 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole, DNA damage-binding protein 1, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028670
To be published
|
|
9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
9FAC
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - mixed polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
6KKZ
 
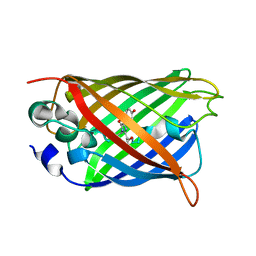 | | Crystal structure of the S65T/F99S/M153T/V163A variant of perdeuterated GFP at pD 8.5 | | Descriptor: | Green fluorescent protein | | Authors: | Tai, Y, Takaba, K, Hanazono, Y, Dao, H.A, Miki, K, Takeda, K. | | Deposit date: | 2019-07-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray crystallographic studies on the hydrogen isotope effects of green fluorescent protein at sub-angstrom resolutions
Acta Crystallogr.,Sect.D, 75, 2019
|
|
9BBE
 
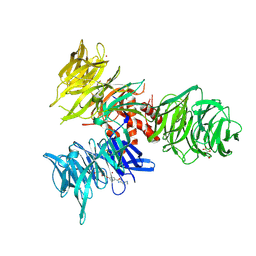 | | Co-crystal structure of human DDB1 bound to fragment UB028668 | | Descriptor: | 5-(4-methoxyphenyl)-3-[(3S)-pyrrolidin-3-yl]-1,2,4-oxadiazole, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028668
To be published
|
|
9BRU
 
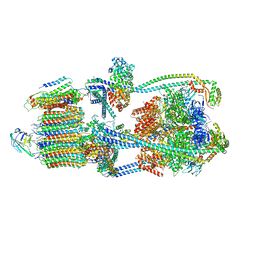 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9FQA
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor PSB-21101 (compound 30b in publication) | | Descriptor: | (5-chloranylpyridin-3-yl) 2-fluoranyl-4-phenylmethoxy-benzoate, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Oneto, A, Guetschow, M, Mueller, C.E. | | Deposit date: | 2024-06-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Nonpeptidic Irreversible Inhibitors of SARS-CoV-2 Main Protease with Potent Antiviral Activity.
J.Med.Chem., 67, 2024
|
|
9GJ2
 
 | | Crystal structure of human cathepsin S produced in insect cells in complex with ketoamide 13b | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Cathepsin S, ... | | Authors: | Falke, S, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Reinke, P.Y.A, Guenther, S, Turk, D, Meents, A. | | Deposit date: | 2024-08-20 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of human cathepsin S produced in insect cells in complex with ketoamide 13b
To Be Published
|
|
9BHN
 
 | |
9JDW
 
 | |
9CYR
 
 | | Crystal structure of D245G mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9F14
 
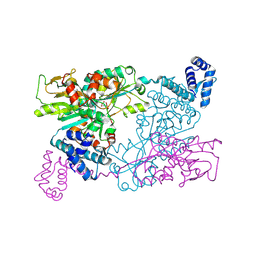 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
9BBG
 
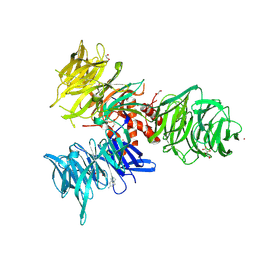 | | Co-crystal structure of human DDB1 bound to fragment UB028671 | | Descriptor: | 1,2-ETHANEDIOL, 1H-indol-6-amine, DNA damage-binding protein 1, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028671
To be published
|
|
9BC3
 
 | | Transglutaminase 2 - Alternate state | | Descriptor: | CALCIUM ION, HB-225 (gluten peptidomimetic TG2 inhibitor), Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Mathews, I.I, Sewa, A.S, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9AT0
 
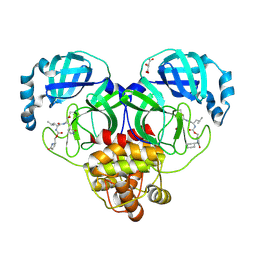 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9BBI
 
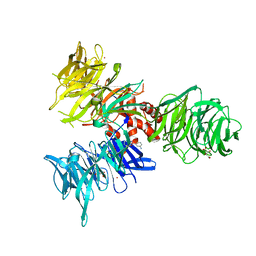 | | Co-crystal structure of human DDB1 bound to fragment UB028669 | | Descriptor: | 3-([1,3]oxazolo[4,5-b]pyridin-2-yl)aniline, DNA damage-binding protein 1, L(+)-TARTARIC ACID, ... | | Authors: | Zeng, H, Dong, A, Frommlet, A, Seitova, A, Loppnau, P, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of human DDB1 bound to fragment UB028669
To be published
|
|
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
8ZN1
 
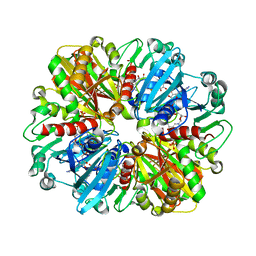 | | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution
To Be Published
|
|
9B1U
 
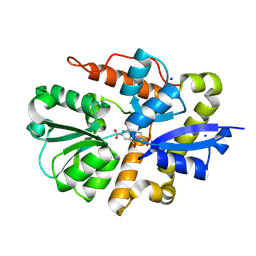 | | Crystal structure of PqqT with PQQ bound | | Descriptor: | PYRROLOQUINOLINE QUINONE, Putative ABC transporter periplasmic solute-binding protein, SODIUM ION | | Authors: | Boggs, D, Bruchs, A, Thompson, P, Olshansky, L, Bridwell-Rabb, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-driven development of a biomimetic rare earth artificial metalloprotein.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9ASW
 
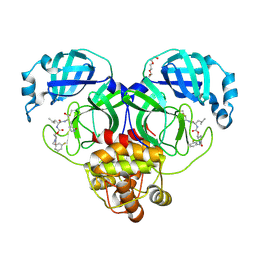 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ENG
 
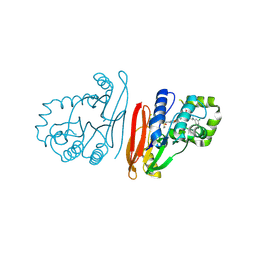 | | Structure of K.pneumoniae LpxH in complex with EBL-3218 | | Descriptor: | MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase, ~{N}-[4-[4-[3,5-bis(chloranyl)phenyl]piperazin-1-yl]sulfonylphenyl]-3-(methylsulfonylamino)benzamide | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2024-03-12 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and in vitro biological evaluation of meta-sulfonamidobenzamide-based antibacterial LpxH inhibitors.
Eur.J.Med.Chem., 278, 2024
|
|
