7OFH
 
 | |
4UTS
 
 | | Room temperature crystal structure of the fast switching M159T mutant of fluorescent protein Dronpa | | Descriptor: | FLUORESCENT PROTEIN DRONPA | | Authors: | Kaucikas, M, Fitzpatrick, A, Bryan, E, Struve, A, Henning, R, Kosheleva, I, Srajer, V, van Thor, J.J. | | Deposit date: | 2014-07-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Room Temperature Crystal Structure of the Fast Switching M159T Mutant of the Fluorescent Protein Dronpa.
Proteins, 83, 2015
|
|
5J62
 
 | | FMN-dependent Nitroreductase (CDR20291_0684) from Clostridium difficile R20291 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, B, Powell, S.M, Hessami, N, Najar, F.Z, Thomas, L.M, West, A.H, Karr, E.A, Richter-Addo, G.B. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole.
Nitric Oxide, 60, 2016
|
|
6Z0C
 
 | | Structure of in silico modelled artificial Maquette-3 protein | | Descriptor: | Maquette-3, POTASSIUM ION | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M.E, Asami, S, Mader, S.L, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of buried charged networks in artificial proteins
Nat Commun, 12, 2021
|
|
4V3O
 
 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | ACETATE ION, CALCIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
6Z35
 
 | | De-novo Maquette 2 protein with buried ion-pair | | Descriptor: | Maquette 2-1ip | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M, Asami, S, Mader, S, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Design of buried charged networks in artificial proteins.
Nat Commun, 12, 2021
|
|
8CE9
 
 | | Crystal structure of human Cd11b I domain in C121 space group | | Descriptor: | 1,2-ETHANEDIOL, Integrin alpha-M, SULFATE ION | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human Cd11b I domain in C121 space group
To Be Published
|
|
8CNX
 
 | | Structure of Enterovirus D68 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of EV D68 3C protease
To Be Published
|
|
6YSJ
 
 | | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-(4-bromophenyl)ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N, Paulus, A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10)
To be published
|
|
1CFW
 
 | | GA-SUBSTITUTED DESULFOREDOXIN | | Descriptor: | GALLIUM (III) ION, PROTEIN (DESULFOREDOXIN), SULFATE ION | | Authors: | Archer, M, Carvalho, A.L, Teixeira, S, Moura, I, Moura, J.J.G, Rusnak, F, Romao, M.J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies by X-ray diffraction on metal substituted desulforedoxin, a rubredoxin-type protein.
Protein Sci., 8, 1999
|
|
1D0X
 
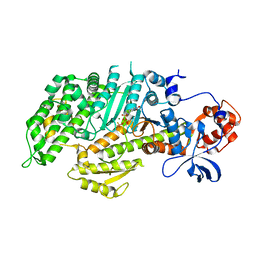 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH M-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | M-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE, MAGNESIUM ION, MYOSIN S1DC MOTOR DOMAIN | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
2ONQ
 
 | |
7NTK
 
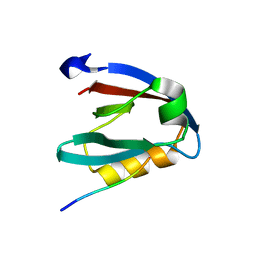 | | PALS1 PDZ1 domain with SARS-CoV-2_E PBM complex | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Envelope small membrane protein, ... | | Authors: | Javorsky, A, Kvansakul, M. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain.
Commun Biol, 4, 2021
|
|
8QEL
 
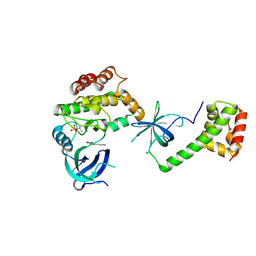 | | PKR kinase domain- eIF2alpha in complex with compound | | Descriptor: | (3~{Z})-3-[(4-methyl-1~{H}-imidazol-5-yl)methylidene]-2-oxidanylidene-1~{H}-indole-5-carboxamide, Eukaryotic translation initiation factor 2 subunit alpha, Interferon-induced, ... | | Authors: | Nawrotek, A, Vuillard, L, Miallau, L. | | Deposit date: | 2023-08-31 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | PKR kinase domain- eIF2alpha in complex with compound
To Be Published
|
|
1D0Y
 
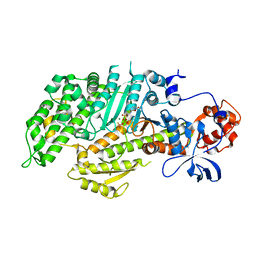 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM FLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN S1DC MOTOR DOMAIN, O-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1CGS
 
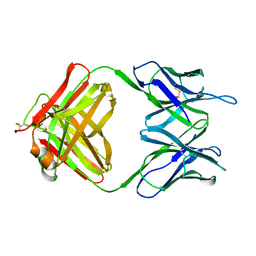 | |
5ITJ
 
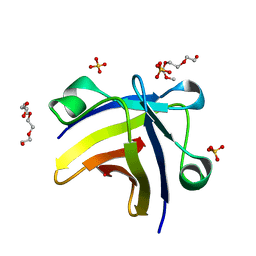 | | The structure of histone-like protein | | Descriptor: | AbrB family transcriptional regulator, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
4V0I
 
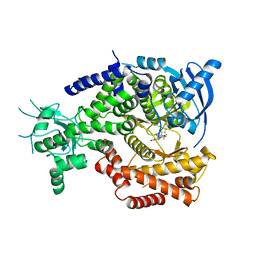 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4UUV
 
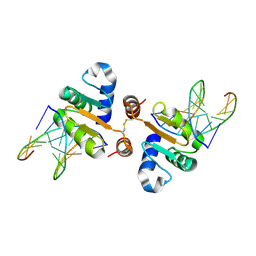 | | Structure of the DNA binding ETS domain of human ETV4 in complex with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP)-3', ETS TRANSLOCATION VARIANT 4 | | Authors: | Newman, J.A, Cooper, C.D.O, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4V11
 
 | | Structure of Synaptotagmin-1 with SV2A peptide phosphorylated at Thr84 | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTIC VESICLE GLYCOPROTEIN 2A, ... | | Authors: | Zhang, N, Gordon, S.L, Fritsch, M.J, Esoof, N, Campbell, D, Gourlay, R, Velupillai, S, Macartney, T, Peggie, M, vanAalten, D.M.F, Cousin, M.A, Alessi, D.R. | | Deposit date: | 2014-09-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of Synaptic Vesicle Protein 2A at Thr84 by Casein Kinase 1 Family Kinases Controls the Specific Retrieval of Synaptotagmin-1.
J.Neurosci., 35, 2015
|
|
4UUL
 
 | |
1D5Z
 
 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | PROTEIN (ENTEROTOXIN TYPE B), PROTEIN (HLA CLASS II HISTOCOMPATIBILITY ANTIGEN), PROTEIN (PEPTIDOMIMETIC INHIBITOR) | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
4V99
 
 | |
5IUF
 
 | |
7O31
 
 | | Crystal structure of the anti-PAS Fab 1.2 in complex with its epitope peptide and the anti-Kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, PAS#1 epitope peptide, anti-Kappa VHH domain, ... | | Authors: | Schilz, J, Schiefner, A, Skerra, A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
