4NZ1
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6HAU
 
 | | KSHV PAN RNA Mta-response element fragment complexed with the globular domain of herpesvirus saimiri ORF57 | | Descriptor: | ACETATE ION, MRE fragment of PAN RNA, ZINC ION, ... | | Authors: | Tunnicliffe, R.B, Levy, C, Ruiz Nivia, H.D, Sandri-Goldin, R.M, Golovanov, A.P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural identification of conserved RNA binding sites in herpesvirus ORF57 homologs: implications for PAN RNA recognition.
Nucleic Acids Res., 47, 2019
|
|
8P6Y
 
 | | Cryo-EM structure of CAK in complex with nucleotide analogue ATPgS | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
6VLJ
 
 | |
5IPZ
 
 | | Crystal structure of human carbonic anhydrase isozyme IV with 5-(2-amino-1,3-thiazol-4-yl)-2-chlorobenzenesulfonamide | | Descriptor: | 5-(2-amino-1,3-thiazol-4-yl)-2-chlorobenzene-1-sulfonamide, Carbonic anhydrase 4, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2016-03-10 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5IQB
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia (CTD of AAC(6')-Ie/APH(2'')-Ia) in complex with GMPPNP, Magnesium, and Kanamycin A | | Descriptor: | Bifunctional AAC/APH, CHLORIDE ION, KANAMYCIN A, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibiotic Binding Drives Catalytic Activation of Aminoglycoside Kinase APH(2)-Ia.
Structure, 24, 2016
|
|
3PJW
 
 | |
5QPW
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000632a | | Descriptor: | 1-methyl-5-(phenylamino)-1,2-dihydro-3H-pyrazol-3-one, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
7S0U
 
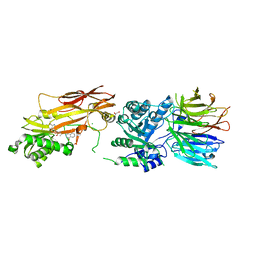 | | PRMT5/MEP50 crystal structure with MTA and phthalazinone fragment bound | | Descriptor: | 1,2-ETHANEDIOL, 4-(aminomethyl)phthalazin-1(2H)-one, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-08-31 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
8F5P
 
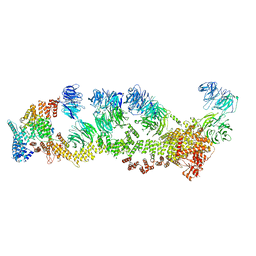 | | Structure of Leishmania tarentolae IFT-A (state 2) | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 122B, putative, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2022-11-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of IFT-A polymerization into trains for ciliary transport.
Cell, 185, 2022
|
|
3NM2
 
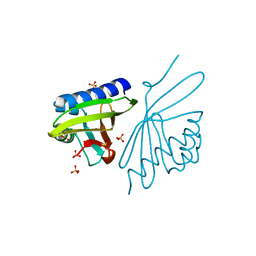 | | Crystal Structure of Ketosteroid Isomerase D38EP39GV40GS42G from Pseudomonas Testosteroni (tKSI) | | Descriptor: | SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38EP39GV40GS42G from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
6CEV
 
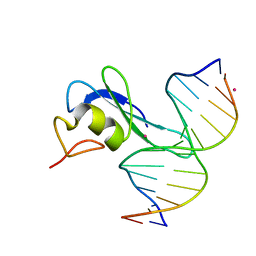 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
5LEL
 
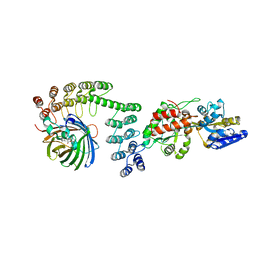 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
4ERQ
 
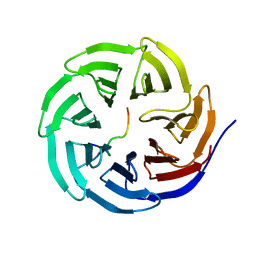 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
6MHW
 
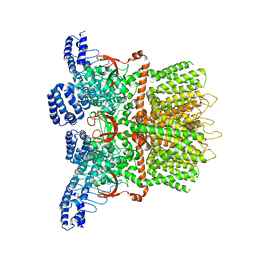 | | Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (1) | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
5LFT
 
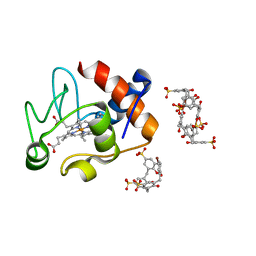 | | Crystal structure of cytochrome c - Bromo-trisulfonatocalix[4]arene complexes | | Descriptor: | BROMIDE ION, Bromo-trisulfonatocalix[4]arene, Cytochrome c iso-1, ... | | Authors: | Doolan, A.M, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2016-07-04 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Protein Recognition by Functionalized Sulfonatocalix[4]arenes.
Chemistry, 24, 2018
|
|
7AMD
 
 | | In situ assembly of choline acetyltransferase ligands by a hydrothiolation reaction reveals key determinants for inhibitor design | | Descriptor: | Choline O-acetyltransferase, SODIUM ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-2,2-dimethyl-4-[[3-[2-[(1~{R})-2-(1-methylpyridin-4-yl)-1-naphthalen-1-yl-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Allgardsson, A, Ekstrom, F.J, Wiktelius, D, Bergstrom, T, Hoster, N, Akfur, C, Forsgren, N, Lejon, C, Hedenstrom, M, Linusson, A. | | Deposit date: | 2020-10-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Situ Assembly of Choline Acetyltransferase Ligands by a Hydrothiolation Reaction Reveals Key Determinants for Inhibitor Design.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5FP0
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, N-cyclopentyl-2-[4-(trifluoromethyl)phenyl]-3H-benzimidazole-4-sulfonamide | | Authors: | Xue, Y, Olsson, T, Johansson, C.A, Oster, L, Beisel, H.G, Rohman, M, Karis, D, Backstrom, S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment Screening of Soluble Epoxide Hydrolase for Lead Generation-Structure-Based Hit Evaluation and Chemistry Exploration.
Chemmedchem, 11, 2016
|
|
5AY8
 
 | | Crystal structure of human nucleosome containing H3.Y | | Descriptor: | CHLORIDE ION, DNA (146-MER), H3.Y, ... | | Authors: | Kujirai, T, Horikoshi, N, Sato, K, Maehara, K, Machida, S, Osakabe, A, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of human histone H3.Y nucleosome
Nucleic Acids Res., 44, 2016
|
|
5IQS
 
 | | WelO5 bound to Fe(II), Cl, and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Mitchell, A.J, Ananth, N, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
4NN1
 
 | |
7AN5
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-10-11 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
7AN6
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-10-11 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
6MLP
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) Y14A mutant from Salmonella typhimurium complexed with histidine | | Descriptor: | ACETATE ION, GLYCEROL, HISTIDINE, ... | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.485 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6RRE
 
 | | SidD, deAMPylase from Legionella pneumophila | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, MAGNESIUM ION | | Authors: | Tascon, I, Lucas, M, Rojas, A.L, Hierro, A. | | Deposit date: | 2019-05-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.586 Å) | | Cite: | Structural insight into the membrane targeting domain of the Legionella deAMPylase SidD.
Plos Pathog., 16, 2020
|
|
