7TGS
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor JOMBt | | Descriptor: | (4S,7R,11S,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
5M7J
 
 | | Blastochloris viridis photosynthetic reaction center structure using best crystal approach | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Sharma, A.S, Johansson, L, Dunevall, E, Wahlgren, W.Y, Neutze, R, Katona, G. | | Deposit date: | 2016-10-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetry in serial femtosecond crystallography data.
Acta Crystallogr A Found Adv, 73, 2017
|
|
6EPU
 
 | | The ATAD2 bromodomain in complex with compound 2 | | Descriptor: | (2~{S})-~{N}-(4-ethanoyl-1,3-thiazol-2-yl)piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
4Y3N
 
 | | Endothiapepsin in complex with fragment 273 | | Descriptor: | 2-[(1S)-2-acetyl-1,2-dihydroisoquinolin-1-yl]-N,N-dimethylacetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
8U7I
 
 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
7U04
 
 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
3WUZ
 
 | | Crystal structure of the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | CITRIC ACID, ISOPROPYL ALCOHOL, Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1QAU
 
 | | UNEXPECTED MODES OF PDZ DOMAIN SCAFFOLDING REVEALED BY STRUCTURE OF NNOS-SYNTROPHIN COMPLEX | | Descriptor: | NEURONAL NITRIC OXIDE SYNTHASE (RESIDUES 1-130) | | Authors: | Hillier, B.J, Christopherson, K.S, Prehoda, K.E, Bredt, D.S, Lim, W.A. | | Deposit date: | 1999-03-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex.
Science, 284, 1999
|
|
2W7O
 
 | | Structure and Activity of Bypass Synthesis by Human DNA Polymerase Kappa Opposite the 7,8-Dihydro-8-oxodeoxyguanosine Adduct | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3', 5'-D(TP*CP*AP*CP*8OGP*GP*AP*AP*TP*CP*CP*TP* TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Egli, M. | | Deposit date: | 2008-12-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural and Functional Elucidation of the Mechanism Promoting Error-Prone Synthesis by Human DNA Polymerase Kappa Opposite the 7,8-Dihydro-8-Oxo-2'-Deoxyguanosine Adduct.
J.Biol.Chem., 284, 2009
|
|
4H8H
 
 | | MUTB inactive double mutant E254Q-D415N | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-09-22 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into product binding in sucrose isomerases from crystal structures of MutB from Rhizobium sp.
To be Published
|
|
4L28
 
 | | Crystal structure of delta516-525 human cystathionine beta-synthase D444N mutant containing C-terminal 6xHis tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-04 | | Release date: | 2013-09-18 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8D0Y
 
 | |
6RZ2
 
 | | SalL with Chloroadenosine | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Adenosyl-chloride synthase | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8CU1
 
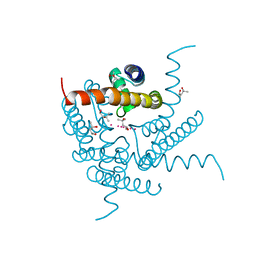 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 500ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU3
 
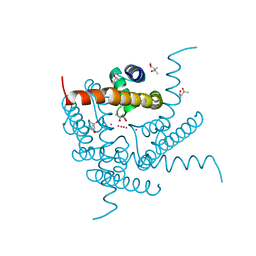 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 200ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU4
 
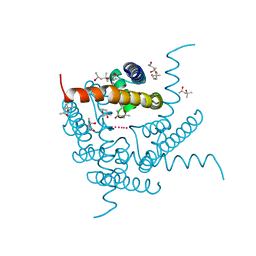 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 1us, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
4Y5J
 
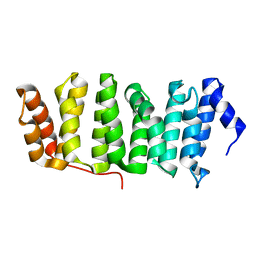 | | Drosophila melanogaster Mini spindles TOG3 | | Descriptor: | MINI SPINDLES TOG3 | | Authors: | Slep, K.C, Howard, A.E. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Drosophila melanogaster Mini Spindles TOG3 Utilizes Unique Structural Elements to Promote Domain Stability and Maintain a TOG1- and TOG2-like Tubulin-binding Surface.
J.Biol.Chem., 290, 2015
|
|
6UHA
 
 | |
6U0U
 
 | | Protofilament Ribbon Flagellar Proteins Rib43a-L | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Khalifa, A.A.Z, Vargas, J, Basu, K, Bui, K.H. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Tubulin lattice in cilia is in a stressed form regulated by microtubule inner proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8TZE
 
 | | Structure of C-terminal half of LRRK2 bound to GZD-824 | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Villagran-Suarez, A, Sanz-Murillo, M, Alegrio-Louro, J, Leschziner, A. | | Deposit date: | 2023-08-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.
Sci Adv, 9, 2023
|
|
4Y63
 
 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-deoxy-5'-{[(2S)-2-(triaza-1,2-dien-2-ium-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
7M8S
 
 | | Crystal Structure of HLA-A*02:01 in complex with KLNDLCFTNV, an 10-mer epitope from SARS-CoV-2 Spike (S386-395) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SARS-CoV-2 Spike-Derived Peptides Presented by HLA Molecules
Biophysica, 1, 2021
|
|
6XHV
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
1QFP
 
 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) | | Descriptor: | PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Burtnick, L, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
4Y3S
 
 | |
