4YBK
 
 | | C-Helix-Out Dasatinib Analog Crystallized with c-Src Kinase | | Descriptor: | 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(4-phenoxyphenyl)-1,3-thiazole-5-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.K, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
2VSP
 
 | | Crystal structure of the fourth PDZ domain of PDZ domain-containing protein 1 | | Descriptor: | PDZ DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Yue, W.W, Shafqat, N, Pilka, E.S, Johansson, C, Murray, J.W, Elkins, J, Roos, A, Cooper, C, Phillips, C, Salah, E, von Delft, F, Doyle, D, Edwards, A, Wikstrom, M, Arrowsmith, C, Bountra, C, Oppermann, U. | | Deposit date: | 2008-04-28 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of the Fourth Pdz Domain of Pdz Domain-Containing Protein 1
To be Published
|
|
5MUW
 
 | | Atomic structure of P4 packaging enzyme fitted into a localized reconstruction of bacteriophage phi6 vertex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
6VC0
 
 | | Crystal structure of the horse MLKL pseudokinase domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
7R5K
 
 | | Human nuclear pore complex (constricted) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science, 376, 2022
|
|
4QOE
 
 | | The value 'crystal structure of fad quinone reductase 2 at 1.45A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The value 'crystal structure of fad quinone reductase 2 at 1.45A
To be Published
|
|
3ON3
 
 | | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA | | Descriptor: | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit, SULFATE ION | | Authors: | Tan, K, Zhang, R, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA
To be Published
|
|
3BD6
 
 | | Glycogen Phosphorylase complex with 1(-D-ribofuranosyl) cyanuric acid | | Descriptor: | 1-beta-D-ribofuranosyl-1,3,5-triazinane-2,4,6-trione, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
7QZD
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikF with an engineered HMA domain of Pikp-1 (Pikp-SNK-EKE) from rice (Oryza sativa) | | Descriptor: | Avr-Pik, Resistance protein Pikp-1 | | Authors: | Maidment, J.H.R, Franceschetti, M, Longya, A, Banfield, M.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effector target-guided engineering of an integrated domain expands the disease resistance profile of a rice NLR immune receptor.
Elife, 12, 2023
|
|
8JOS
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | Descriptor: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
3ZM1
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
8Q8I
 
 | | AO75L in complex with a synthetic trisaccharide acceptor. | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-methyl-6-octoxy-3-oxidanyl-oxan-4-yl]oxy-6-methyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, BICINE, ... | | Authors: | Laugueri, M.E, Speciale, I, Gimeno, A, Sicheng, L, Poveda, A, Lowary, T, Van Etten J, L, Barbero, J, De Castro, C, Tonetti, M, Rojas A, L. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | AO75L in complex with a synthetic trisaccharide acceptor.
To Be Published
|
|
7US3
 
 | | Structure of Putrescine N-hydroxylase Involved Complexed with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putrescine N-hydroxylase, ... | | Authors: | Tanner, J.J, Bogner, A.N. | | Deposit date: | 2022-04-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N -Hydroxylase from Acinetobacter baumannii.
Biochemistry, 61, 2022
|
|
4DKY
 
 | | Crystal structure Analysis of N terminal region containing the dimerization domain and DNA binding domain of HU protein(Histone like protein-DNA binding) from Mycobacterium tuberculosis [H37Ra] | | Descriptor: | DNA-binding protein HU homolog, MANGANESE (II) ION | | Authors: | Bhowmick, T, Ramagopal, U.A, Ghosh, S, Nagaraja, V, Ramakumar, S. | | Deposit date: | 2012-02-05 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors
Nat Commun, 5, 2014
|
|
5QQB
 
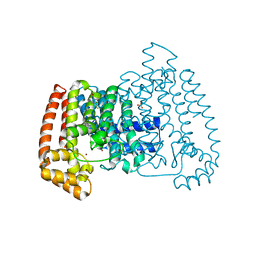 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000676a | | Descriptor: | 8-fluoranyl-5-methyl-1,2,3,6-tetrahydro-1,5-benzodiazocin-4-one, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
6BRM
 
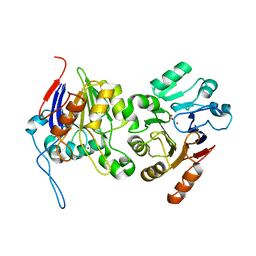 | | The crystal structure of isothiocyanate hydrolase from Delia radicum gut bacteria | | Descriptor: | FORMIC ACID, Putative metal-dependent isothiocyanate hydrolase SaxA, ZINC ION | | Authors: | Tan, K, van den Bosch, T, Joachimiak, A, Welte, C. | | Deposit date: | 2017-11-30 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Functional Profiling and Crystal Structures of Isothiocyanate Hydrolases Found in Gut-Associated and Plant-Pathogenic Bacteria.
Appl. Environ. Microbiol., 84, 2018
|
|
6ZOK
 
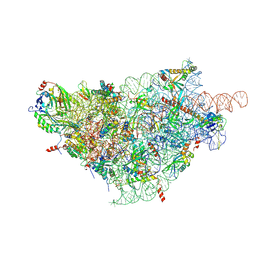 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V9T
 
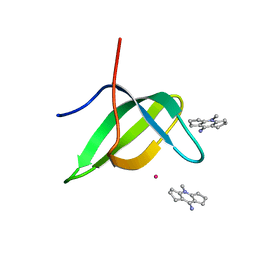 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Tudor domain-containing protein 3, UNKNOWN ATOM OR ION | | Authors: | Li, W, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
1W4F
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
6Y0G
 
 | | Structure of human ribosome in classical-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
5MO2
 
 | | Neutron structure of cationic trypsin in complex with N-amidinopiperidine | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
6BYJ
 
 | | Structure of human 14-3-3 gamma bound to O-GlcNAc peptide | | Descriptor: | 14-3-3 protein gamma, 2-acetamido-2-deoxy-beta-D-glucopyranose, TSTTATPPVSQASSTTTSTW O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
4YCG
 
 | | Pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, Bone Morphogenetic Protein 9 Prodomain, ... | | Authors: | Mi, L.-Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6H3O
 
 | | Alcohol oxidase from Phanerochaete chrysosporium mutant F101S | | Descriptor: | Alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Nguyen, Q.-T, Romero, E, Dijkman, W.P, de Vasconcellos, S.P, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2018-07-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Engineering of Phanerochaete chrysosporium Alcohol Oxidase for Enhanced Oxidative Power toward Glycerol.
Biochemistry, 57, 2018
|
|
5JGV
 
 | | Spin-Labeled T4 Lysozyme Construct A73V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
