6YAL
 
 | | Mammalian 48S late-stage initiation complex with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S Ribosomal protein uS3, 40S Ribosomal protein uS7, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
2EG2
 
 | | The crystal structure of PII protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Nitrogen regulatory protein P-II | | Authors: | Sakai, H, Shinkai, A, Kitamura, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of PII protein
To be Published
|
|
6SED
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose | | Descriptor: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
8R0Q
 
 | |
5C3T
 
 | | PD-1 binding domain from human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
8R0W
 
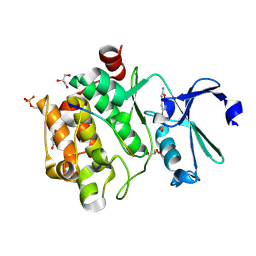 | |
8R1W
 
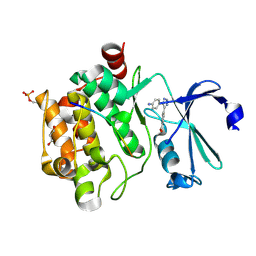 | | Pim1 in complex with 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid and Pimtide | | Descriptor: | 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pim1 in complex with 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid and Pimtide
To Be Published
|
|
5IN2
 
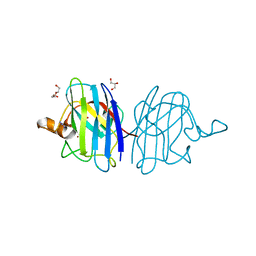 | |
5G4G
 
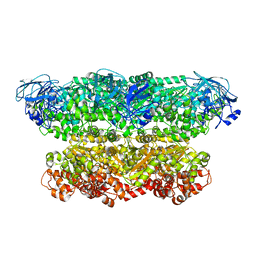 | | Structure of the ATPgS-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5MYF
 
 | | Convergent evolution involving dimeric and trimeric dUTPases in signalling. | | Descriptor: | dUTPase from DI S. aureus phage | | Authors: | Donderis, J, Bowring, J, Maiques, E, Ciges-Tomas, J.R, Alite, C, Mehmedov, I, Tormo-Mas, M.A, Penades, J.R, Marina, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization.
PLoS Pathog., 13, 2017
|
|
2WXG
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with SW13. | | Descriptor: | 2-{[4-amino-3-(3-fluoro-5-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
6OPE
 
 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to near-cognate 70S A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Sunita, S, Dunham, C.M. | | Deposit date: | 2019-04-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YNW
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - central stalk/cring | | Descriptor: | subunit c, subunit delta, subunit epsilon, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
8QDF
 
 | | Engineered LmrR with Met-89 replaced by para-boronophenylalanine | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | Deposit date: | 2023-08-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Boron catalysis in a designer enzyme.
Nature, 629, 2024
|
|
5MTU
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
6IGP
 
 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
7QZ9
 
 | |
6OSK
 
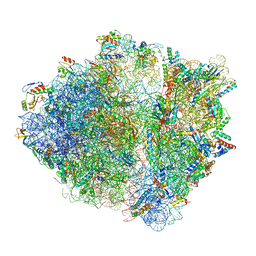 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
7ALM
 
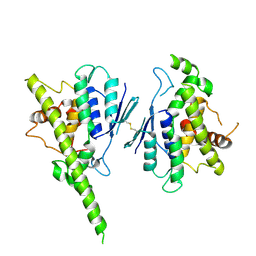 | |
7QZ6
 
 | |
5IPU
 
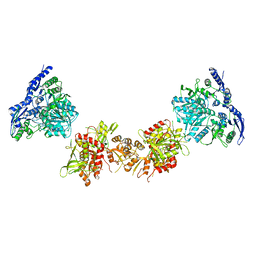 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 6 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
6OVO
 
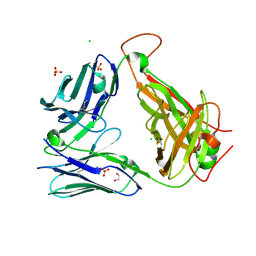 | | Crystal structure of the unliganded PG10 TCR | | Descriptor: | 1,2-ETHANEDIOL, Alpha Chain T-Cell Receptor PG10, Beta Chain T-Cell Receptor PG10, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A TCR beta-Chain Motif Biases toward Recognition of Human CD1 Proteins.
J Immunol., 203, 2019
|
|
3D0O
 
 | |
5IPN
 
 | | SigmaS-transcription initiation complex with 4-nt nascent RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2016-03-09 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.61 Å) | | Cite: | Structures of E. coli sigma S-transcription initiation complexes provide new insights into polymerase mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
