6HOK
 
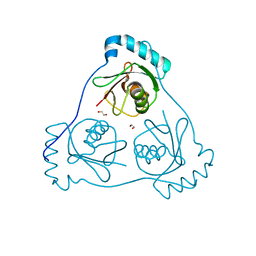 | | Structure of Beclin1 LIR (S96E) motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
5WH1
 
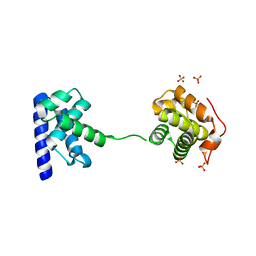 | |
8Q4P
 
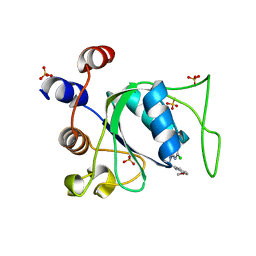 | |
8Q2R
 
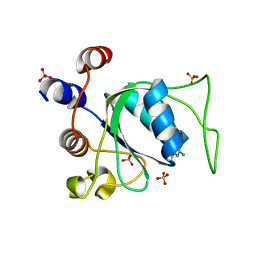 | | Crystal structure of YTHDC1 in complex with Compound 3 (ZA_431) | | Descriptor: | 5-chloranyl-~{N},3-dimethyl-1~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Li, Y, Caflisch, A. | | Deposit date: | 2023-08-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
4XBD
 
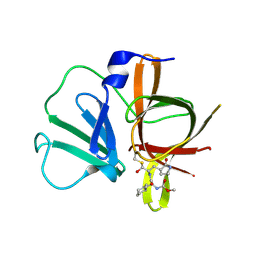 | | 1.45A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Orthorhombic P Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
8Q2Y
 
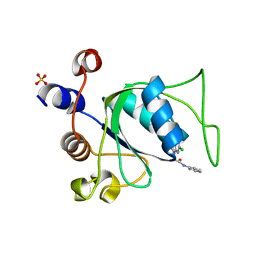 | |
6HP5
 
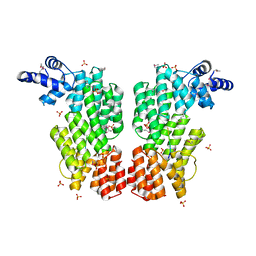 | | ARBITRIUM PEPTIDE RECEPTOR FROM SPBETA PHAGE | | Descriptor: | GLY-MET-PRO-ARG-GLY-ALA, SPBc2 prophage-derived uncharacterized protein YopK, SULFATE ION | | Authors: | Marina, A, Gallego del Sol, F. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems.
Mol.Cell, 74, 2019
|
|
8FEJ
 
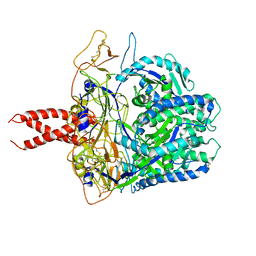 | |
6QN4
 
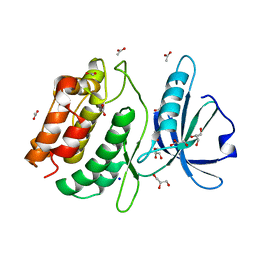 | |
6DIC
 
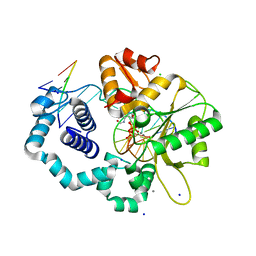 | | D276G DNA polymerase beta substrate complex with templating cytosine and incoming Fapy-dGTP analog | | Descriptor: | 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
8TA2
 
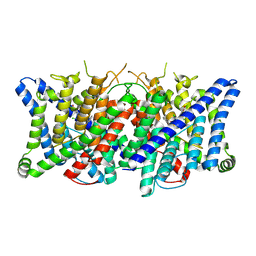 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8Q3G
 
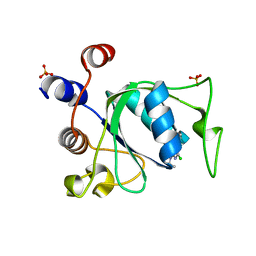 | |
8THE
 
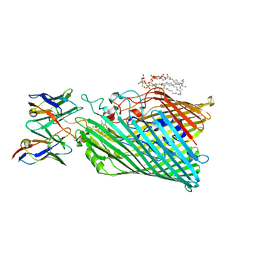 | | Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme | | Descriptor: | (2~{R},4~{R},5~{S},6~{S})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{S})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Heme/hemoglobin uptake outer membrane receptor PhuR, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Knejski, P, Erramilli, S.K, Kossiakoff, A.A. | | Deposit date: | 2023-07-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Chaperone-assisted cryo-EM structure of P. aeruginosa PhuR reveals molecular basis for heme binding.
Structure, 32, 2024
|
|
8Q4W
 
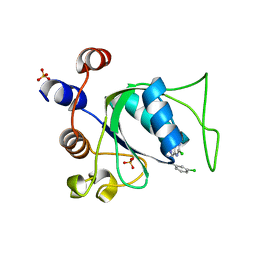 | | Crystal structure of YTHDC1 in complex with Compound 40 (CS_3a) | | Descriptor: | 5-chloranyl-3-[(3-chlorophenyl)methyl]-~{N}-methyl-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Caflisch, A. | | Deposit date: | 2023-08-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
8Q2X
 
 | |
6HQ4
 
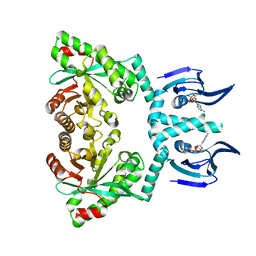 | | Structure of EAL enzyme Bd1971 - cAMP bound form | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, EAL Enzyme Bd1971, MAGNESIUM ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
6U5H
 
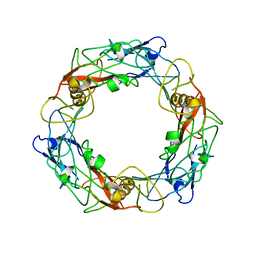 | | CryoEM Structure of Pyocin R2 - precontracted - hub | | Descriptor: | Probable bacteriophage protein Pyocin R2 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
8Q2S
 
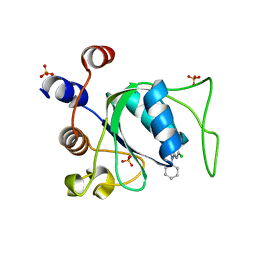 | |
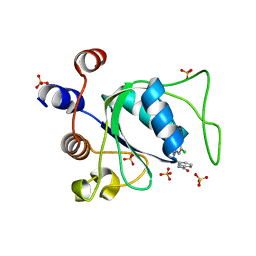
8Q3A
 
 | |
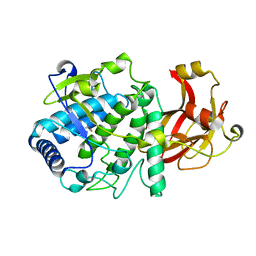
6HQJ
 
 | |
8Q35
 
 | |
6QV0
 
 | | Structure of ATP-bound outward-facing TM287/288 in complex with sybody Sb_TM35 | | Descriptor: | ABC transporter, ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hutter, C.A.J, Huerlimann, L.M, Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The extracellular gate shapes the energy profile of an ABC exporter.
Nat Commun, 10, 2019
|
|
8Q4N
 
 | |
6DJB
 
 | | Structure of human Volume Regulated Anion Channel composed of SWELL1 (LRRC8A) | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kefauver, J.M, Saotome, K, Pallesen, J, Cottrell, C.A, Ward, A.B, Patapoutian, A. | | Deposit date: | 2018-05-24 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the human volume regulated anion channel.
Elife, 7, 2018
|
|
6QVO
 
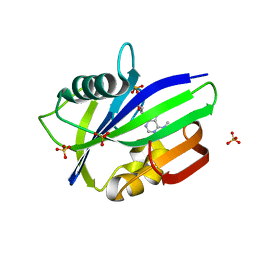 | | Crystal structure of human MTH1 in complex with N6-methyl-dAMP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, N6-METHYL-DEOXY-ADENOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Scaletti, E, Vallin, K.S, Brautigam, L, Sarno, A, Warpman Berglund, U, Helleday, T, Stenmark, P, Jemth, A.S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | MutT homologue 1 (MTH1) removes N6-methyl-dATP from the dNTP pool.
J.Biol.Chem., 295, 2020
|
|
