8VU8
 
 | |
6E68
 
 | | NAMPT co-crystal with inhibitor compound 2 | | Descriptor: | (2E)-N-{4-[1-(3-aminobenzene-1-carbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Waight, A.B, Neumann, C.S. | | Deposit date: | 2018-07-24 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NAMPT co-crystal with inhibitor compound 2
to be published
|
|
2XJ4
 
 | |
8CK1
 
 | | Carin 1 bacteriophage tail, connector and tail fibers assembly | | Descriptor: | Connector Protein, Tail Nozzle, Tail fibers Dpo36 | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
6RG4
 
 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzyl-4-methylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(3~{S})-4-methyl-3-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
8V9O
 
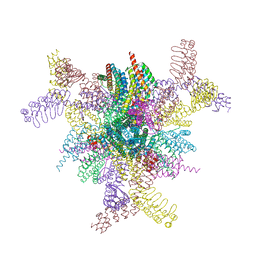 | | Imaging scaffold engineered to bind the therapeutic protein target BARD1 | | Descriptor: | CALCIUM ION, Tetrahedral Nanocage Cage Component Fused to Anti-BARD1 Darpin, Tetrahedral Nanocage Cage, ... | | Authors: | Agdanowski, M.P, Castells-Graells, R, Sawaya, M.R, Yeates, T.O, Arbing, M.A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | X-ray crystal structure of a designed rigidified imaging scaffold in the ligand-free conformation.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6EBR
 
 | |
7KVF
 
 | |
6W4S
 
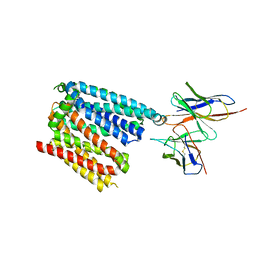 | | Structure of apo human ferroportin in lipid nanodisc | | Descriptor: | Fab45D8 Heavy Chain, Fab45D8 Light Chain, Solute carrier family 40 member 1 | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
2HO5
 
 | | Crystal structure of Oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae | | Descriptor: | Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of Oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae
To be Published
|
|
6EAX
 
 | | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21 | | Descriptor: | CALCIUM ION, CYS-THR-LYS-SER-ILE-CYS, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.189 Å) | | Cite: | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21
To Be Published
|
|
4WVC
 
 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
5YSG
 
 | |
4WEV
 
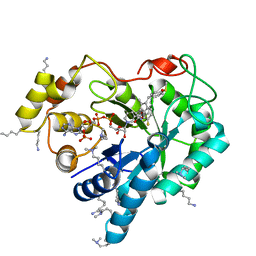 | | Crystal structure of human AKR1B10 complexed with NADP+ and sulindac | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Crespo, I, Porte, S, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10.
Chem.Biol.Interact., 234, 2015
|
|
4X94
 
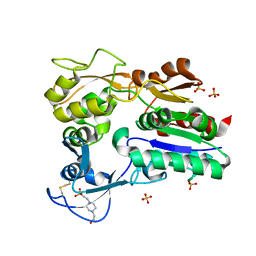 | |
8V47
 
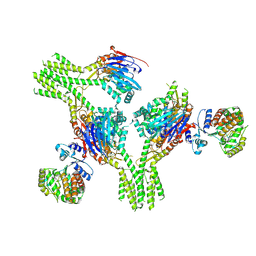 | | CryoEM structure of AriA-AriB complex (Form II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
3AU0
 
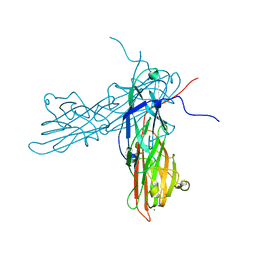 | | Structural and biochemical characterization of ClfB:ligand interactions | | Descriptor: | Clumping factor B, MAGNESIUM ION | | Authors: | Ganesh, V.K, Barbu, E.M, Deivanayagam, C.C.S, Le, B, Anderson, A.S, Matsuka, Y, Lin, S.L, Foster, T.F, Narayana, S.V.L, Hook, M. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biochemical characterization of ClfB:ligand interactions
To be published
|
|
8V46
 
 | | CryoEM structure of AriA-AriB complex (Form I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB, ... | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
7S3T
 
 | | NzeB Diketopiperazine Dimerase Mutant: Q68I-G87A-A89G-I90V | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Harris, N.R, Shende, V.V, Sanders, J.N, Newmister, S.A, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 2023
|
|
6RRK
 
 | | Crystal structure of the central region of human cohesin subunit STAG1 in complex with RAD21 peptide | | Descriptor: | Cohesin subunit SA-1, Double-strand-break repair protein rad21 homolog | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6CB0
 
 | | Crystal Structure of the FAK FERM domain | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Dementiev, A, Marlowe, T. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High resolution crystal structure of the FAK FERM domain reveals new insights on the Druggability of tyrosine 397 and the Src SH3 binding site.
BMC Mol Cell Biol, 20, 2019
|
|
5NLV
 
 | | Brag2 Sec7-PH (390-763) | | Descriptor: | IQ motif and SEC7 domain-containing protein 1 | | Authors: | Nawrotek, A, Cherfils, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7NT1
 
 | | Crystal structure of SARS CoV2 main protease in complex with FSP007 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [(2R)-1-[2-(1H-indol-3-yl)ethylamino]-1-oxidanylidene-butan-2-yl] prop-2-enoate | | Authors: | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6VIU
 
 | |
