8AY8
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-iSB9-HAB1 TERNARY COMPLEX | | Descriptor: | Abscisic acid receptor PYL1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8AYA
 
 | |
5NM8
 
 | | Structure of PipY, the COG0325 family member of Synechococcus elongatus PCC7942, with PLP bound | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, PipY | | Authors: | Tremino, L, Forcada-Nadal, A, Contreras, A, Rubio, V. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Studies on cyanobacterial protein PipY shed light on structure, potential functions, and vitamin B6 -dependent epilepsy.
FEBS Lett., 591, 2017
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | Descriptor: | rRNA expansion segment ES27L in an "out" conformation | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4Z02
 
 | | Crystal structure of BRD1 in complex with Isoquinoline-3-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, UNKNOWN ATOM OR ION, ... | | Authors: | DONG, A, IQBAL, A, WALKER, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of BRD1 incomplex with Isoquinoline-3-carboxylic acid
to be published
|
|
8AY9
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-ABA-HAB1 TERNARY COMPLEX | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL1, CHLORIDE ION, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
5KLN
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169A in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
8AY7
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-iSB7-HAB1 TERNARY COMPLEX | | Descriptor: | Abscisic acid receptor PYL1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.131 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
4Z0Z
 
 | |
6G2U
 
 | | Crystal structure of the human glutamate dehydrogenase 2 (hGDH2) | | Descriptor: | CHLORIDE ION, Glutamate dehydrogenase 2, mitochondrial, ... | | Authors: | Fadouloglou, V.F, Dimovasili, C, Providaki, M, Kotsifaki, D, Sarrou, I, Plaitakis, A, Zaganas, I, Kokkinidis, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.934287 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 2, a positively selected novel human enzyme involved in brain biology and cancer pathophysiology.
J.Neurochem., 2021
|
|
6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6G3T
 
 | | X-ray structure of NSD3-PWWP1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-26 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
5KLK
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ and 2-hydroxymuconate-6-semialdehyde | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
6GJ8
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH BI 2852 | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GNT
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-(6-pyrrolidin-1-ylquinolin-5-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GOJ
 
 | | X-ray structure of the adduct formed upon reaction of lysozyme with a Pt(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated triphenylphosphonium cation | | Descriptor: | CHLORIDE ION, Lysozyme C, NITRATE ION, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
6T98
 
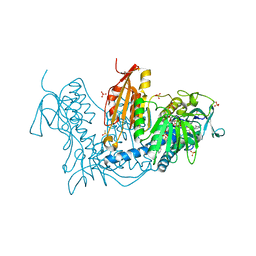 | | Trypanothione Reductase from Leishmania infantum in complex with 9a | | Descriptor: | 4-[3-methyl-1-[4-[4-(2-phenylethyl)-1,3-thiazol-2-yl]-3-(2-piperidin-4-ylethoxy)phenyl]-1,2,3-triazol-3-ium-4-yl]butan-1-amine, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
4ZE3
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
4YZ2
 
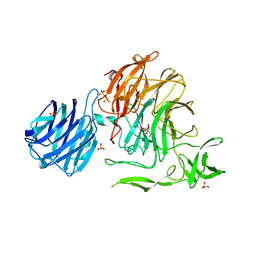 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SULFATE ION, Sialidase NanC | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YZV
 
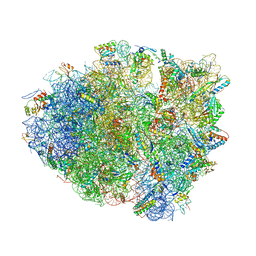 | | Precleavage 70S structure of the P. vulgaris HigB deltaH92 toxin bound to the ACA codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5L84
 
 | |
4YZ1
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, apo structure. | | Descriptor: | Putative neuraminidase, SULFATE ION | | Authors: | Lukacik, P, Owen, D.O, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
6GZE
 
 | | Tubulin-GDP.BeF complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Oliva, M.A, Diaz, J.F. | | Deposit date: | 2018-07-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural model for differential cap maturation at growing microtubule ends.
Elife, 9, 2020
|
|
4ZFQ
 
 | | Structure of M. tuberculosis (3,3) L,D-Transpeptidase, LdtMt5. (Meropenen-adduct form) | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, DI(HYDROXYETHYL)ETHER, L,D-transpeptidase 5 | | Authors: | Basta, L, Ghosh, A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-04-21 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
5LBP
 
 | | Oceanobacillus iheyensis macrodomain mutant N30A | | Descriptor: | MacroD-type macrodomain, PHOSPHATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
