6XKS
 
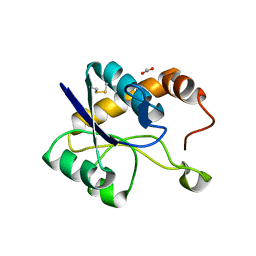 | | Crystal structure of domain A from the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) of Salmonella typhimurium | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Histidine ABC transporter substrate-binding protein HisJ | | Authors: | Romero-Romero, S, Berrocal, T, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermodynamic and kinetic analysis of the LAO binding protein and its isolated domains reveal non-additivity in stability, folding and function.
Febs J., 2023
|
|
6XQ6
 
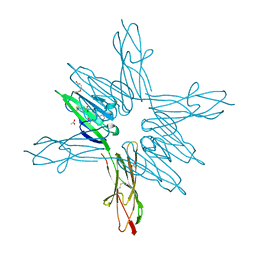 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 11 | | Descriptor: | 3-phenoxyphenol, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ3
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-(3-{[3-(4-carboxyphenoxy)phenyl]methoxy}phenyl)-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ8
 
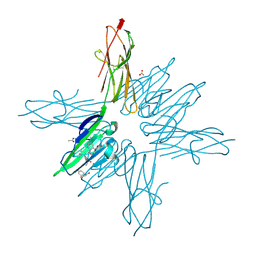 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 11 | | Descriptor: | 3-phenoxyphenol, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ7
 
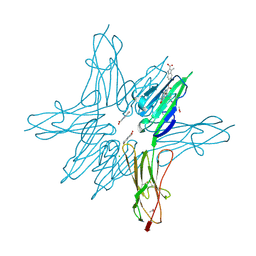 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 5 | | Descriptor: | 5-bromo-3-methyl-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
4UOR
 
 | | Structure of lipoteichoic acid synthase LtaS from Listeria monocytogenes in complex with glycerol phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, LIPOTEICHOIC ACID SYNTHASE, MAGNESIUM ION | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and mechanistic insight into the Listeria monocytogenes two-enzyme lipoteichoic acid synthesis system.
J. Biol. Chem., 289, 2014
|
|
6XQ1
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-[3-({[3-(4-carboxyphenoxy)phenyl]methoxy}methyl)phenyl]-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
7PXC
 
 | | Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state A) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Kavalchuk, M, Weber-Ban, E. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex.
Nat Commun, 13, 2022
|
|
6XQV
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | Descriptor: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
4UIK
 
 | | crystal structure of quinine-dependent Fab 314.1 | | Descriptor: | FAB 314.1 | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
4UM1
 
 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3573 | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
4UQ6
 
 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
5MC6
 
 | | Cryo-EM structure of a native ribosome-Ski2-Ski3-Ski8 complex from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Schmidt, C, Kowalinski, E, Shanmuganathan, V, Defenouillere, Q, Braunger, K, Heuer, A, Pech, M, Namane, A, Berninghausen, O, Fromont-Racine, M, Jacquier, A, Conti, E, Becker, T, Beckmann, R. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex.
Science, 354, 2016
|
|
4UOP
 
 | | Crystal structure of the lipoteichoic acid synthase LtaP from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, LIPOTEICHOIC ACID PRIMASE, MAGNESIUM ION, ... | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-06-06 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic insight into the Listeria monocytogenes two-enzyme lipoteichoic acid synthesis system.
J. Biol. Chem., 289, 2014
|
|
7Q5B
 
 | | Cryo-EM structure of Ty3 retrotransposon targeting a TFIIIB-bound tRNA gene | | Descriptor: | DNA (19-MER), DNA (31-MER), DNA (34-MER), ... | | Authors: | Abascal-Palacios, G, Jochem, L, Pla-Prats, C, Beuron, F, Vannini, A. | | Deposit date: | 2021-11-03 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis of Ty3 retrotransposon integration at RNA Polymerase III-transcribed genes.
Nat Commun, 12, 2021
|
|
5MEI
 
 | | Crystal structure of Agelastatin A bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | McClary, B, Zinshteyn, B, Meyer, M, Jouanneau, M, Pellegrino, S, Yusupova, G, Schuller, A, Reyes, J.C.P, Lu, J, Luo, C, Dang, Y, Romo, D, Yusupov, M, Green, R, Liu, J.O. | | Deposit date: | 2016-11-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A.
Cell Chem Biol, 24, 2017
|
|
4UOO
 
 | |
6Y8D
 
 | | 14-3-3 Sigma in complex with phosphorylated caspase{pS164} peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4UIN
 
 | | crystal structure of quinine-dependent Fab 314.3 with quinine | | Descriptor: | FAB 314.3, Quinine | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
7PE1
 
 | |
7PE2
 
 | |
4UFN
 
 | | Laboratory evolved variant R-C1B1 of potato epoxide hydrolase StEH1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, EPOXIDE HYDROLASE | | Authors: | Carlsson, A.J, Bauer, P, Nilsson, M, Dobritzsch, D, Kamerlin, S.C.L, Widersten, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Diversity and Enantioconvergence in Potato Epoxide Hydrolase 1.
Org.Biomol.Chem., 14, 2016
|
|
4UCP
 
 | |
6Y6D
 
 | | Tubulin-7-Aminonoscapine complex | | Descriptor: | (3~{S})-7-azanyl-6-methoxy-3-[(5~{R})-4-methoxy-6-methyl-7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-3~{H}-2-benzofuran-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Prota, A.E, Rodriguez-Salarichs, J, Gu, W, Bennani, Y.L, Jimenez-Barbero, J, Canales, A, Steinmetz, M.O, Diaz, J.F. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Noscapine Activation for Tubulin Binding.
J.Med.Chem., 63, 2020
|
|
4UCN
 
 | |
