2G3B
 
 | | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | GLYCEROL, putative TetR-family transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Wang, S, Koclega, K.D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
3KXS
 
 | |
6DM0
 
 | | Open state GluA2 in complex with STZ and blocked by IEM-1460, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
4V9N
 
 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
2G4B
 
 | | Structure of U2AF65 variant with polyuridine tract | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', Splicing factor U2AF 65 kDa subunit | | Authors: | Sickmier, E.A, Kielkopf, C.L. | | Deposit date: | 2006-02-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of polypyrimidine tract recognition
by the essential splicing factor U2AF65.
Mol.Cell, 23, 2006
|
|
8SVB
 
 | | Antimicrobial lasso peptide achromonodin-1 | | Descriptor: | Achromonodin-1 | | Authors: | Carson, D.V, Cheung-Lee, W.L, So, L, Link, A.J. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Bioactivity of the Achromonodins: Lasso Peptides Encoded by Achromobacter .
J.Nat.Prod., 86, 2023
|
|
7M4F
 
 | | DNA Polymerase Lambda, dCTP:At Mg2+ Product State Ternary Complex, 300 min | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
5L78
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in NAD+ bound form) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Pena, I.A, Rembeza, E, Strain-Damerell, C, Chalk, R, Borkowska, O, Goubin, S, Velupillai, S, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Arruda, P, Yue, W.W. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in NAD+ bound form)
To Be Published
|
|
5L7S
 
 | |
4V8J
 
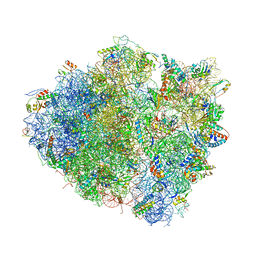 | | Crystal structure of the bacterial ribosome ram mutation G347U. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2011-12-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7M4G
 
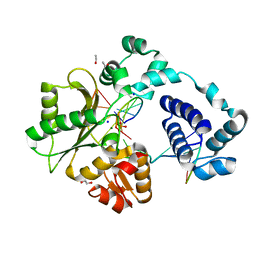 | | DNA Polymerase Lambda, dCTP:At Mg2+ Product State Ternary Complex, 960 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7QZC
 
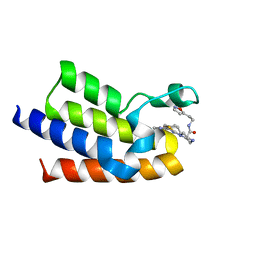 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 104 | | Descriptor: | (2R)-1-[4-(4-ethanoyl-3-ethyl-5-methyl-1H-pyrrol-2-yl)-1,3-thiazol-2-yl]-N-[2-(1,2-oxazol-5-yl)ethyl]piperazine-2-carboxamide, Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-31 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7M4I
 
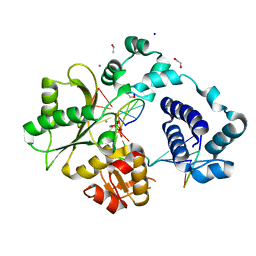 | | DNA Polymerase Lambda, dCTP:At Mn2+ Product State Ternary Complex, 420 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7R01
 
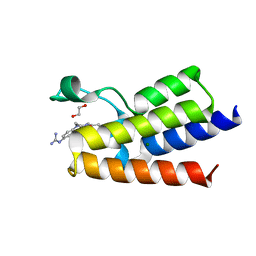 | | BAZ2A bromodomain in complex with N-acetylpiperazine derivative fragment 18 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(4-ethanoylpiperazin-1-yl)phenyl]guanidine, Bromodomain adjacent to zinc finger domain protein 2A, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
4WCE
 
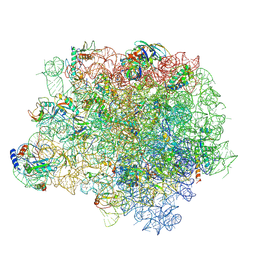 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-04 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6DVI
 
 | |
4USF
 
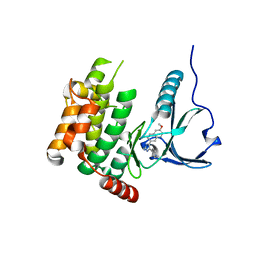 | | Human SLK with SB-440719 | | Descriptor: | 4-[4-(6-methoxynaphthalen-2-yl)-1H-imidazol-5-yl]pyridine, STE20-LIKE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
2GA9
 
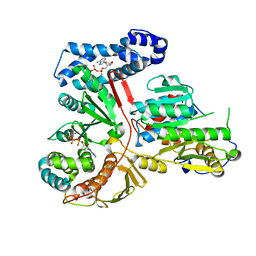 | | Crystal Structure of the Heterodimeric Vaccinia Virus Polyadenylate Polymerase with Bound ATP-gamma-S | | Descriptor: | CALCIUM ION, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Moure, C.M, Bowman, B.R, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2006-03-08 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity.
Mol.Cell, 22, 2006
|
|
7MB1
 
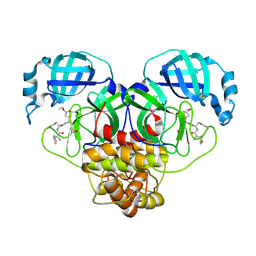 | |
3KMF
 
 | |
5W3J
 
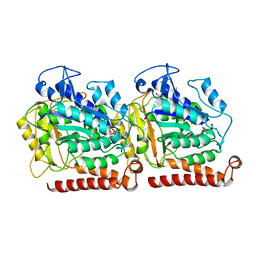 | | Yeast microtubule stabilized with Taxol assembled from mutated tubulin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
4UX7
 
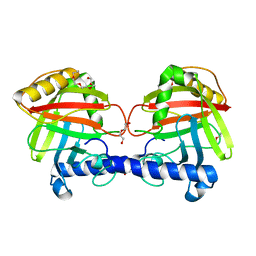 | | Structure of a Clostridium difficile sortase | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PUTATIVE PEPTIDASE C60B, ... | | Authors: | Chambers, C.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2014-08-19 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Function of a Clostridium Difficile Sortase Enzyme.
Sci.Rep., 5, 2015
|
|
4UY9
 
 | | Structure of MLK1 kinase domain with leucine zipper 1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 9 | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
4UZ8
 
 | | The SeMet structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B), SULFATE ION | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2015-05-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
1WBE
 
 | | X-ray structure of bovine GLTP | | Descriptor: | DECANOIC ACID, GLYCEROL, GLYCOLIPID TRANSFER PROTEIN | | Authors: | Airenne, T.T, Kidron, H, West, G, Nymalm, Y, Nylund, M, Mattjus, P, Salminen, T.A. | | Deposit date: | 2004-11-01 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Evidence for Adaptive Ligand Binding of Glycolipid Transfer Protein.
J.Mol.Biol., 355, 2006
|
|
