5KH7
 
 | | Crystal structure of fragment (3-[6-Oxo-3-(3-pyridinyl)-1(6H)-pyridazinyl]propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(6-oxidanylidene-3-pyridin-3-yl-pyridazin-1-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Walker, J, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
3H5Z
 
 | | Crystal Structure of Leishmania major N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Qiu, W, Hutchinson, A, Lin, Y.-H, Wernimont, A, Mackenzie, F, Ravichandran, M, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Fairlamb, A.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | N-myristoyltransferase inhibitors as new leads to treat sleeping sickness.
Nature, 464, 2010
|
|
6T9F
 
 | | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.24847341 Å) | | Cite: | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
4QJ5
 
 | | Structure of a fragment of human phospholipase C-beta3 delta472-581, bound to IP3 and in complex with Galphaq | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Molecular mechanisms of phospholipase C beta 3 autoinhibition.
Structure, 22, 2014
|
|
5DJZ
 
 | | Fc Heterodimer Design 7.8 D399M/Y407A + T366V/K409V | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-02 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
1AE2
 
 | | MUTANT L32R OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
6TB0
 
 | | Crystal structure of thermostable omega transaminase 4-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
6PSF
 
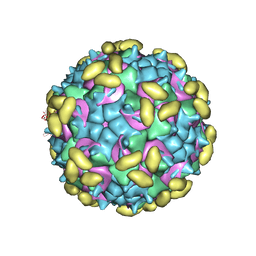 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | Descriptor: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5UPY
 
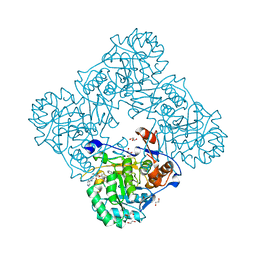 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21
To Be Published
|
|
6TNC
 
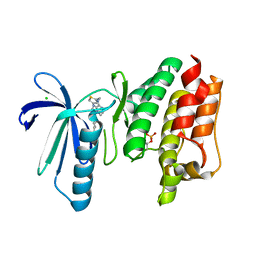 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 46 | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Marquardt, T, Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
4QLT
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 2 (PR924) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4ZKV
 
 | |
6TOP
 
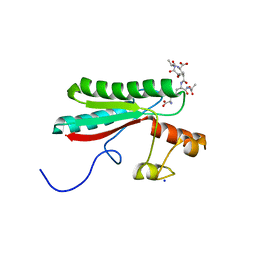 | | Structure of the PorE C-terminal domain, a protein of T9SS from Porphyromonas gingivalis | | Descriptor: | GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, OmpA family protein, SODIUM ION | | Authors: | Trinh, T.N, Cambillau, C, Roussel, A, Leone, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Type IX secretion system PorE C-terminal domain from Porphyromonas gingivalis in complex with a peptidoglycan fragment.
Sci Rep, 10, 2020
|
|
8WOP
 
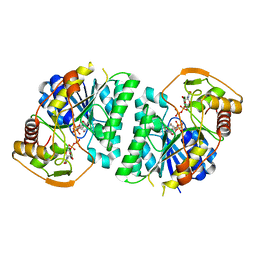 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, wild-type | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
6D62
 
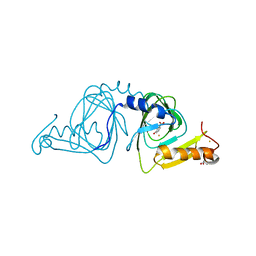 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6TPI
 
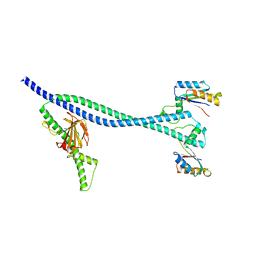 | | EnvC bound to the FtsX periplasmic domain | | Descriptor: | Cell division protein FtsX, Murein hydrolase activator EnvC | | Authors: | Crow, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into bacterial cell division from a structure of EnvC bound to the FtsX periplasmic domain.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6D4K
 
 | |
5UXQ
 
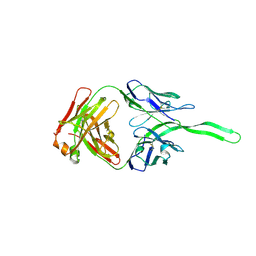 | | Structure of anti-HIV trimer apex antibody PGT143 | | Descriptor: | PGT143 Fab Heavy Chain, PGT143 Fab Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic Beta-Hairpin Structure
Immunity, 46, 2017
|
|
4Z86
 
 | |
7LBE
 
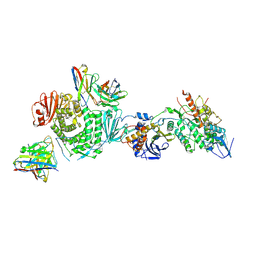 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
8JTZ
 
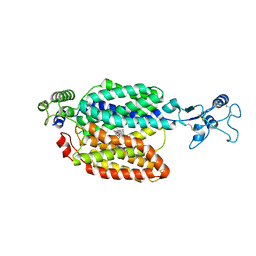 | | hOCT1 in complex with spironolactone in outward facing partially occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
6TEI
 
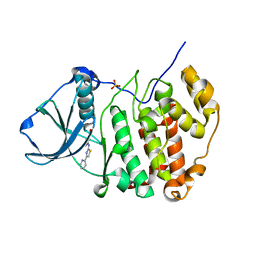 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the 2-aminothiazole-type inhibitor 17 | | Descriptor: | 3-[(4-pyridin-2-yl-1,3-thiazol-2-yl)amino]benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
8JTX
 
 | | hOCT1 in complex with nb5660 in inward facing fully open conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
5VC9
 
 | | Zinc finger of human CXXC4 in complex with CpG DNA | | Descriptor: | CXXC-type zinc finger protein 4, CpG DNA, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
8A31
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC694 | | Descriptor: | 4-(3-fluoranylpyrrol-1-yl)-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
