3ERN
 
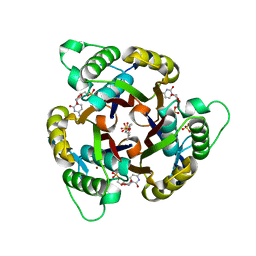 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with AraCMP | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTOSINE ARABINOSE-5'-PHOSPHATE, GERANYL DIPHOSPHATE, ... | | Authors: | Hunter, W.N, Ramsden, N.L, Kemp, L.A. | | Deposit date: | 2008-10-02 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
2ICI
 
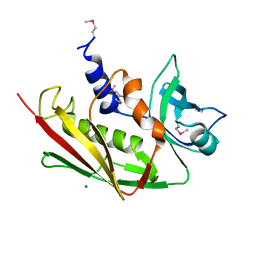 | |
5KLS
 
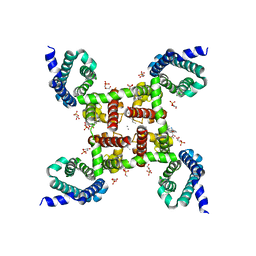 | | Structure of CavAb in complex with Br-dihydropyridine derivative UK-59811 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
8IKD
 
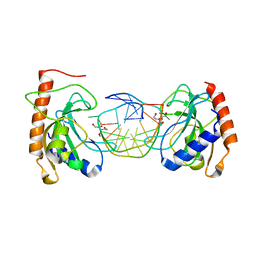 | |
1KAK
 
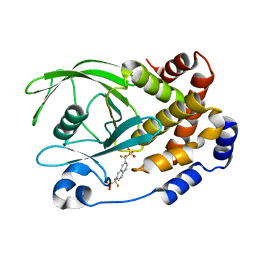 | | Human Tyrosine Phosphatase 1B Complexed with an Inhibitor | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, {[7-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALEN-2-YL]-DIFLUORO-METHYL}-PHOSPHONIC ACID | | Authors: | Jia, Z, Ye, Q, Dinaut, A.N, Wang, Q, Waddleton, D, Payette, P, Ramachandran, C, Kennedy, B, Hum, G, Taylor, S.D. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of protein tyrosine phosphatase 1B in complex with inhibitors bearing two phosphotyrosine mimetics.
J.Med.Chem., 44, 2001
|
|
1SXX
 
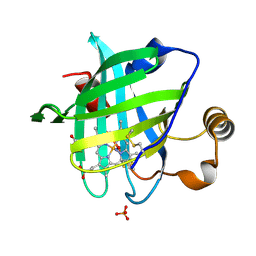 | | 1.0 A Crystal Structure of D129A/L130A Mutant of Nitrophorin 4 Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
7T7O
 
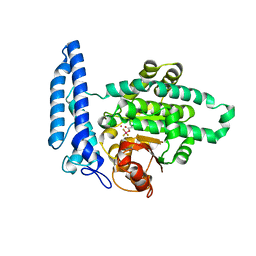 | |
5KMH
 
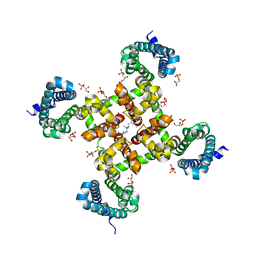 | | Structure of CavAb in complex with Br-verapamil | | Descriptor: | (2~{R})-2-(2-bromophenyl)-5-[2-(3,4-dimethoxyphenyl)ethyl-methyl-amino]-2-propan-2-yl-pentanenitrile, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
3ESY
 
 | | E16KE61K Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
8A45
 
 | |
8A5K
 
 | |
4RJF
 
 | |
6JGR
 
 | | Crystal structure of barley exohydrolaseI W434Y in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
1T8G
 
 | | Crystal structure of phage T4 lysozyme mutant L32A/L33A/T34A/C54T/C97A/E108V | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | He, M.M, Wood, Z.A, Baase, W.A, Xiao, H, Matthews, B.W. | | Deposit date: | 2004-05-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation.
Protein Sci., 13, 2004
|
|
6QA9
 
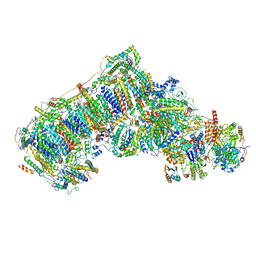 | | Isolated complex I class refinement from Ovine respiratory supercomplex I+III2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-19 | | Release date: | 2019-08-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QC8
 
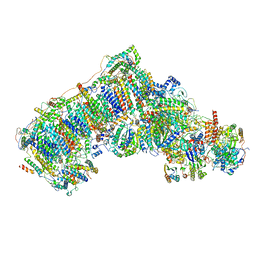 | | Ovine respiratory complex I FRC open class 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6PYY
 
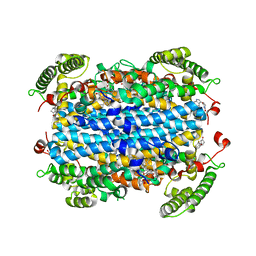 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PF-06840003 in Active Site and Exo site | | Descriptor: | (3S)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Pham, K.N, Lewis-Ballester, A, Yeh, S.R. | | Deposit date: | 2019-07-31 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in Human Indoleamine 2,3-Dioxygenase 1 and Tryptophan Dioxygenase.
J.Am.Chem.Soc., 141, 2019
|
|
4RJ0
 
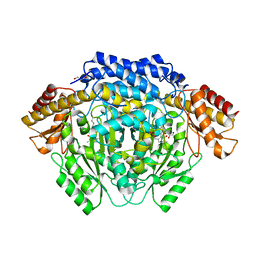 | |
4RIZ
 
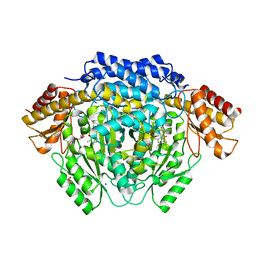 | |
8A7V
 
 | |
7TA6
 
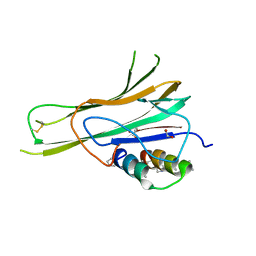 | | Trimer-to-Monomer Disruption of Tumor Necrosis Factor-alpha (TNF-alpha) by unnatural alpha/beta-peptide-1 | | Descriptor: | 1,2-ETHANEDIOL, AMINO GROUP, Alpha/Beta-peptide-1, ... | | Authors: | Niu, J, Bingman, C.A, Gellman, S.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Trimer-to-Monomer Disruption Mechanism for a Potent, Protease-Resistant Antagonist of Tumor Necrosis Factor-alpha Signaling.
J.Am.Chem.Soc., 144, 2022
|
|
1TAH
 
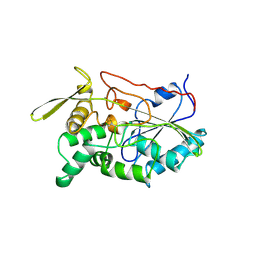 | | THE CRYSTAL STRUCTURE OF TRIACYLGLYCEROL LIPASE FROM PSEUDOMONAS GLUMAE REVEALS A PARTIALLY REDUNDANT CATALYTIC ASPARTATE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Noble, M.E.M, Cleasby, A, Johnson, L.N, Egmond, M, Frenken, L.G.J. | | Deposit date: | 1993-12-21 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate.
FEBS Lett., 331, 1993
|
|
4RKV
 
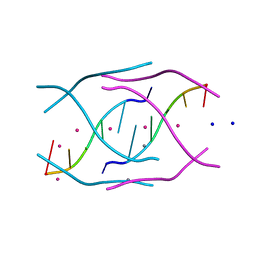 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
7TGS
 
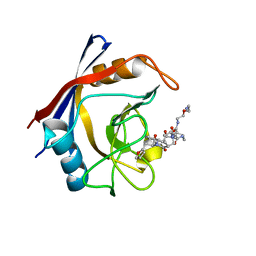 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor JOMBt | | Descriptor: | (4S,7R,11S,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
5KZV
 
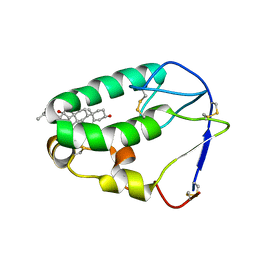 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in complex with 20(S)-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Smoothened | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.616 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
