6GHU
 
 | |
8D2P
 
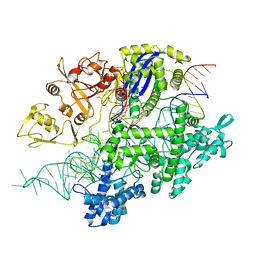 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA target strand (5'-D(P*CP*CP*AP*GP*GP*AP*TP*CP*TP*TP*GP*CP*CP*AP*TP*CP*CP*TP*AP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8OZ9
 
 | |
5V11
 
 | |
8P3C
 
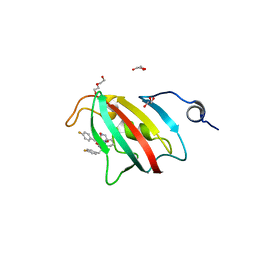 | | Full length structure of BpMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8P3D
 
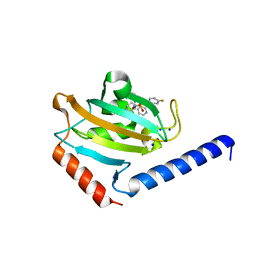 | | Full length structure of TcMIP with bound inhibitor NJS224. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-4-methyl-1-oxidanylidene-1-(pyridin-3-ylmethylamino)pentan-2-yl]piperidine-2-carboxamide, SODIUM ION, peptidylprolyl isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
6FGW
 
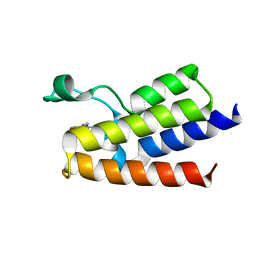 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 4 | | Descriptor: | 1-methyl-6-oxidanylidene-~{N}-(2-pyrrolidin-1-ylethyl)pyridine-3-carboxamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
5CYB
 
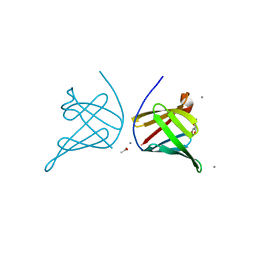 | | Structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Abdullah, M.R, Hammerschmidt, S, Hermoso, J.A. | | Deposit date: | 2015-07-30 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the function and structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae
To Be Published
|
|
5ETE
 
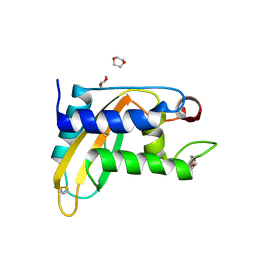 | |
4RUR
 
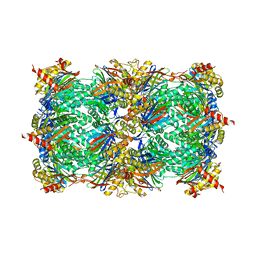 | | Yeast 20S proteasome in complex with the alkaloid indolo-phakellin (4) | | Descriptor: | (2E,3aR,14aS)-9-bromo-2-imino-1,2,3,5,6,14a-hexahydro-4H,8H-imidazo[4',5':5,6]pyrrolo[1',2':4,5]pyrazino[1,2-a]indol-8-one, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Beck, P, Lansdell, T.A, Hewlett, N.M, Tepe, J.J, Groll, M. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Indolo-Phakellins as beta 5-Specific Noncovalent Proteasome Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4YV7
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL | | Descriptor: | GLYCEROL, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL
To be published
|
|
3K24
 
 | | Crystal structure of mature apo-Cathepsin L C25A mutant in complex with Gln-Leu-Ala peptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-6)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin L1, ... | | Authors: | Adams-Cioaba, M.A, Krupa, J.C, Mort, J.S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition and cleavage of histone H3 by cathepsin L.
Nat Commun, 2, 2011
|
|
5HW6
 
 | |
5D7C
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
6BTW
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a phenyl-pteridinone inhibitor | | Descriptor: | 2-[(3,5-difluoro-4-hydroxyphenyl)amino]-8-phenyl-7,8-dihydropteridin-6(5H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a phenyl-pteridinone inhibitor
To Be Published
|
|
5HW0
 
 | |
5V42
 
 | |
3JPP
 
 | | Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-MonoMethyl Methylene triphosphate | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy({(S)-hydroxy[(1R)-1-phosphonoethyl]phosphoryl}oxy)phosphoryl]guanosine, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Batra, V.K, Upton, J, Kashmerov, B, Beard, W.A, Wilson, S.H, Goodman, M.F, McKenna, C.E. | | Deposit date: | 2009-09-04 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Halogenated beta,gamma-Methylene- and Ethylidene-dGTP-DNA Ternary Complexes with DNA Polymerase beta: Structural Evidence for Stereospecific Binding of the Fluoromethylene Analogues.
J.Am.Chem.Soc., 132, 2010
|
|
7KP5
 
 | |
5V4L
 
 | |
5UUV
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with a product IMP and the inhibitor P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with a product IMP and the inhibitor P182
To Be Published
|
|
6C9R
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(thiophen-3-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | 9-beta-D-ribofuranosyl-6-(thiophen-3-yl)-9H-purine, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
4JI8
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.742 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
3JR6
 
 | |
4TKO
 
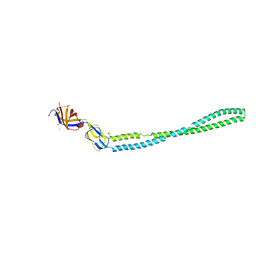 | | Structure of the periplasmic adaptor protein EmrA | | Descriptor: | EmrA, ISOPROPYL ALCOHOL, MAGNESIUM ION | | Authors: | Hinchliffe, P, Greene, N.P, Paterson, N.G, Crow, A, Hughes, C, Koronakis, V. | | Deposit date: | 2014-05-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump.
Febs Lett., 588, 2014
|
|
