7K4W
 
 | | Crystal structure of Kemp Eliminase HG3.17 in the inactive state | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
6GX1
 
 | |
7Q4Y
 
 | |
8F0W
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
7K8M
 
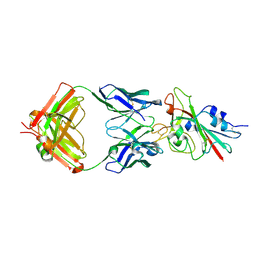 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
6FUJ
 
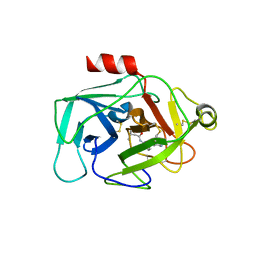 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | Descriptor: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
7K8V
 
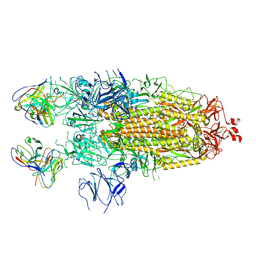 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C110 Fab Heavy Chain, ... | | Authors: | Dam, K.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
4FAU
 
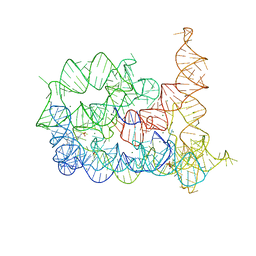 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+ and 5'-exon | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Group IIC intron, MAGNESIUM ION, ... | | Authors: | Marcia, M, Pyle, A.M. | | Deposit date: | 2012-05-22 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Visualizing Group II Intron Catalysis through the Stages of Splicing.
Cell(Cambridge,Mass.), 151, 2012
|
|
6VU8
 
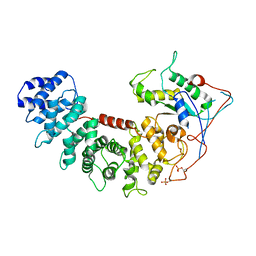 | | Structure of G-alpha-i bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
6FRJ
 
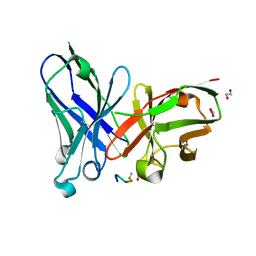 | | Crystal structure of scFv-SM3 in complex with APD-SeThrGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, APD-SeThr-RP, ... | | Authors: | Companon, I, Castro-Lopez, J, Escudero-Casao, M, Avenoza, A, Busto, J.H, Castillon, S, Jimenez-Barbero, J, Bernardes, G.J, Boutureira, O, Jimenez-Oses, G, Asensio, J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2018-02-16 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Design of Potent Tumor-Associated Antigens: Modulation of Peptide Presentation by Single-Atom O/S or O/Se Substitutions at the Glycosidic Linkage.
J. Am. Chem. Soc., 141, 2019
|
|
5JV0
 
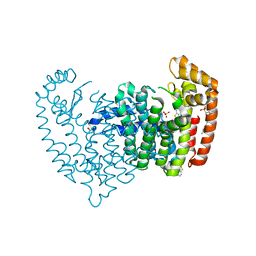 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-08-038 | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION, [(1R)-2-(3-fluorophenyl)-1-{[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}ethyl]phosphonic acid | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
6BXK
 
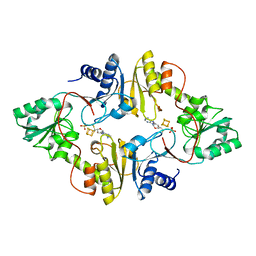 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and MTA | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
7JV1
 
 | | Crystal Structure of KSR1:MEK1 in complex with AMP-PNP, and allosteric MEK inhibitor APS-9-95-1 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 1, MAGNESIUM ION, ... | | Authors: | Khan, Z.M, Dar, A.C, Scopton, A.P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural basis for the action of the drug trametinib at KSR-bound MEK.
Nature, 588, 2020
|
|
7K8O
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C002 | | Descriptor: | C002 Fab Heavy Chain, C002 Fab Light Chain, GLYCEROL, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
4S0O
 
 | |
5V7P
 
 | | Atomic structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase), in complex with a monobody | | Descriptor: | DECANE, Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-03-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
8OP2
 
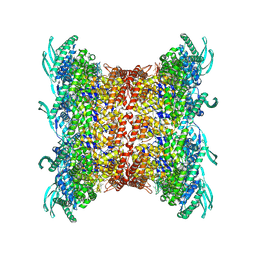 | |
6C5C
 
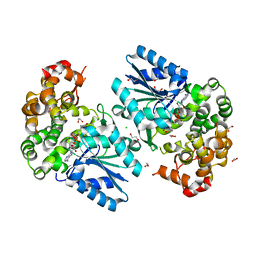 | | Crystal structure of the 3-dehydroquinate synthase (DHQS) domain of Aro1 from Candida albicans SC5314 in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, CHLORIDE ION, ... | | Authors: | Michalska, K, Evdokimova, E, Di Leo, R, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
8PTL
 
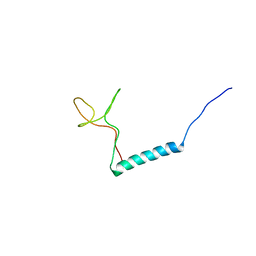 | | human PHOX2B C-terminal domain including the polyA fragment at 278K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
5HYJ
 
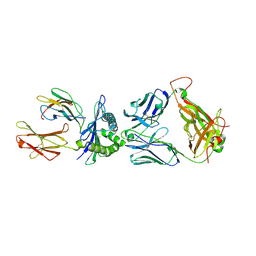 | | 1E6 TCR in Complex with HLA-A02 carrying AQWGPDPAAA | | Descriptor: | ALA-GLN-TRP-GLY-PRO-ASP-PRO-ALA-ALA-ALA, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2016-02-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A "hotspot" for autoimmune T cells in type 1 diabetes.
J.Clin.Invest., 126, 2016
|
|
4ZCE
 
 | | Crystal Structure of the dust mite allergen Der p 23 from Dermatophagoides pteronyssinus | | Descriptor: | 1,2-ETHANEDIOL, Dust mite allergen | | Authors: | Pedersen, L.C, Mueller, G.A, Randall, T.A, Glesner, J, Perera, L, Edwards, L.L, Chapman, M.D, London, R.E, Pomes, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serological, genomic and structural analyses of the major mite allergen Der p 23.
Clin Exp Allergy, 46, 2016
|
|
5Y33
 
 | |
4ZDV
 
 | |
5H6S
 
 | | Crystal structure of Hydrazidase S179A mutant complexed with a substrate | | Descriptor: | 4-oxidanylbenzohydrazide, Amidase | | Authors: | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
8OOU
 
 | |
