8D41
 
 | | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254 | | Descriptor: | (1R,2R,3S,4S)-3-[4-(4-fluorophenyl)piperazine-1-carbonyl]bicyclo[2.2.1]heptane-2-carboxylic acid, Coatomer subunit beta', GLYCEROL, ... | | Authors: | Zeng, H, Saraon, P, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254
To Be Published
|
|
6B0L
 
 | | KLP10A-AMPPNP in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
7SR4
 
 | |
1TTL
 
 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1YS2
 
 | | Burkholderia cepacia lipase complexed with hexylphosphonic acid (S) 2-methyl-3-phenylpropyl ester | | Descriptor: | CALCIUM ION, HEXYLPHOSPHONIC ACID (S)-2-METHYL-3-PHENYLPROPYL ESTER, Lipase | | Authors: | Mezzetti, A, Schrag, J.D, Cheong, C.S, Kazlauskas, R.J. | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mirror-Image Packing in Enantiomer Discrimination Molecular Basis for the Enantioselectivity of B.cepacia Lipase toward 2-Methyl-3-Phenyl-1-Propanol.
Chem.Biol., 12, 2005
|
|
8TBP
 
 | | HLA-DRB1*15:01 in complex with smith antigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ting, Y.T, Broury, A, Ooi, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.12621117 Å) | | Cite: | Smith-specific regulatory T cells halt the progression of lupus nephritis.
Nat Commun, 15, 2024
|
|
6WJO
 
 | | Crystal structure of wild-type Arginine Repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to tyrosine | | Descriptor: | Arginine repressor, SODIUM ION, SULFATE ION, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6DST
 
 | |
1YTP
 
 | |
4XIC
 
 | | ANTPHD WITH 15BP di-thioate modified DNA DUPLEX | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C2S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
6AZZ
 
 | | Crystal structure of Pfs25 in complex with the transmission blocking antibody 1190 | | Descriptor: | 1190 antibody, heavy chain, light chain, ... | | Authors: | Scally, S.W, McLeod, B, Bosch, A, King, C.R, Julien, J.P. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular definition of multiple sites of antibody inhibition of malaria transmission-blocking vaccine antigen Pfs25.
Nat Commun, 8, 2017
|
|
6R7R
 
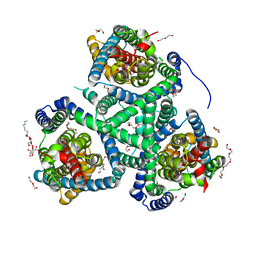 | | Crystal structure of the glutamate transporter homologue GltTk in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, DECYL-BETA-D-MALTOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arkhipova, V, Guskov, A, Slotboom, D.J. | | Deposit date: | 2019-03-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and transport of D-aspartate by the glutamate transporter homolog Glt Tk .
Elife, 8, 2019
|
|
6DYF
 
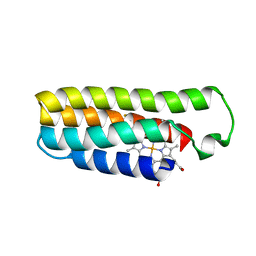 | | Cu(II)-bound structure of the engineered cyt cb562 variant, CH3Y | | Descriptor: | CHLORIDE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
5IF6
 
 | |
6DYH
 
 | |
7SR0
 
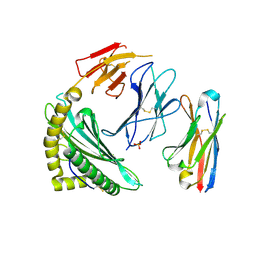 | | Single chain trimer HLA-A*02:01 (H98L, Y108C) with HPV.16 E7 peptide YMLDLQPET | | Descriptor: | PHOSPHATE ION, Protein E7 peptide,Beta-2-microglobulin,MHC class I antigen chimera, VHH | | Authors: | Finton, K.A.K, Rupert, P.B. | | Deposit date: | 2021-11-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Effects of HLA single chain trimer design on peptide presentation and stability.
Front Immunol, 14, 2023
|
|
6ANN
 
 | |
4H7V
 
 | |
7SR3
 
 | |
1U3B
 
 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
8DEL
 
 | |
3IU5
 
 | | Crystal structure of the first bromodomain of human poly-bromodomain containing protein 1 (PB1) | | Descriptor: | Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-30 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
830C
 
 | | COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID | | Descriptor: | 4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYLMETHYL]-TETRAHYDRO-PYRAN-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
8CZI
 
 | |
5IIZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F0 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
