7JMJ
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 37 - State 5 (S5) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
8V7Q
 
 | |
6CAJ
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B from Homo sapiens | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Tsai, J.C, Miller-Vedam, L.E, Anand, A.A, Jaishankar, P, Nguyen, H.C, Renslo, A.R, Frost, A, Walter, P. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule.
Science, 359, 2018
|
|
5F5R
 
 | | TRAP1N-ADPNP | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, MAGNESIUM ION, ... | | Authors: | Tsai, F.T.F, Lee, S, Sung, N, Lee, J, Chang, C, Joachimiak, A. | | Deposit date: | 2015-12-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8A2R
 
 | | Cryo-EM structure of F-actin in the Mg2+-ADP-BeF3- nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
6GF1
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | SULFATE ION, Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
7QVE
 
 | | Spinach 20S proteasome | | Descriptor: | Proteasome subunit alpha type, Proteasome subunit alpha type-3, Proteasome subunit beta, ... | | Authors: | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the plant 26S proteasome.
Plant Commun., 3, 2022
|
|
5KSX
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor AM-02-072 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [[(2~{S})-2-[[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoyl]amino]phosphonic acid | | Authors: | Park, J, Matralis, A, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
8RE0
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
4RSU
 
 | | Crystal structure of the light and hvem complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, W, Ramagoal, U.A, Himmel, D, Bonanno, J.B, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
8V5E
 
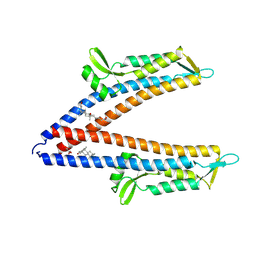 | | IpaD (122-321) Pi-helix Mutant (delta Q148) Bound to Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J. | | Deposit date: | 2023-11-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 2024
|
|
6FH3
 
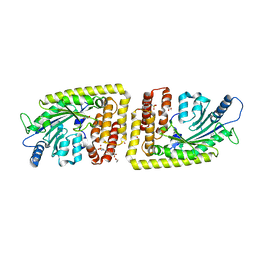 | | Protein arginine kinase McsB in the pArg-bound state | | Descriptor: | 1,2-ETHANEDIOL, Protein-arginine kinase, phospho-arginine | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
8DHU
 
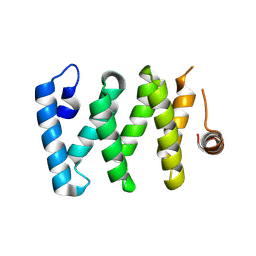 | | Crystal structure of LARP-DM15 from Drosophila melanogaster bound to m7GpppC | | Descriptor: | La-related protein 1, MAGNESIUM ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-9-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[(2~{R},3~{S},4~{R},5~{R})-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Nguyen, E, Berman, A.J. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Comparative analysis of the LARP1 C-terminal DM15 region through Coelomate evolution.
Plos One, 19, 2024
|
|
8DIO
 
 | |
5KU5
 
 | |
7K5E
 
 | | 1.75 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor JAG-5-7 | | Descriptor: | 4-{[(5-fluoro-2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
6GH2
 
 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-glc-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
8OYD
 
 | |
7KPZ
 
 | |
8RG1
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - wild type pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
6GI4
 
 | |
6FJP
 
 | | Adenovirus species 26 knob protein, high resolution, High pH | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Biorxiv, 2019
|
|
1M90
 
 | | Co-crystal structure of CCA-Phe-caproic acid-biotin and sparsomycin bound to the 50S ribosomal subunit | | Descriptor: | 23S RRNA, 5S RRNA, 6-AMINOHEXANOIC ACID, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into peptide bond formation.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8VRJ
 
 | | Rigid body fitted model for gamma tubulin ring complex capped microtubule | | Descriptor: | Actin, cytoplasmic 1, Gamma-tubulin complex component 3, ... | | Authors: | Aher, A, Urnavicius, L, Kapoor, T.M. | | Deposit date: | 2024-01-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the gamma-tubulin ring complex-capped microtubule.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VJB
 
 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
