6WCO
 
 | |
7QRW
 
 | |
6SPG
 
 | | Pseudomonas aeruginosa 70s ribosome from a clinical isolate | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | 登録日 | 2019-09-01 | | 公開日 | 2019-10-16 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6X0K
 
 | |
6XKL
 
 | | SARS-CoV-2 HexaPro S One RBD up | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wrapp, D, Hsieh, C.-L, Goldsmith, J.A, McLellan, J.S. | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-15 | | 最終更新日 | 2020-09-30 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structure-based design of prefusion-stabilized SARS-CoV-2 spikes.
Science, 369, 2020
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
8FYV
 
 | | Salmonella enterica serovar Typhimurium chemoreceptor Tsr (taxis to serine and repellents) ligand-binding domain in complex with l-serine | | 分子名称: | CHLORIDE ION, Methyl-accepting chemotaxis protein, SERINE, ... | | 著者 | Baylink, A, Gentry-Lear, Z, Glenn, S. | | 登録日 | 2023-01-26 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bacterial vampirism mediated through taxis to serum.
Elife, 12, 2024
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
6WKP
 
 | | Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | 著者 | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-16 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6X5H
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP | | 分子名称: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | 著者 | Kadirvelraj, R, Wood, Z.A. | | 登録日 | 2020-05-26 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
6QDH
 
 | |
5NNP
 
 | | Structure of Naa15/Naa10 bound to HypK-THB | | 分子名称: | CARBOXYMETHYL COENZYME *A, GLYCEROL, N-terminal acetyltransferase-like protein, ... | | 著者 | Weyer, F.A, Gumiero, A, Kopp, J, Sinning, I. | | 登録日 | 2017-04-10 | | 公開日 | 2017-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | Structural basis of HypK regulating N-terminal acetylation by the NatA complex.
Nat Commun, 8, 2017
|
|
8FK0
 
 | |
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
6XHX
 
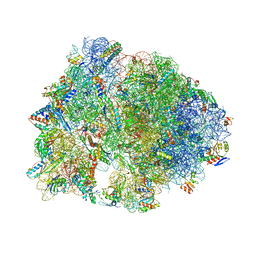 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with erythromycin and protein Y (YfiA) at 2.55A resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | 登録日 | 2020-06-19 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
6WN9
 
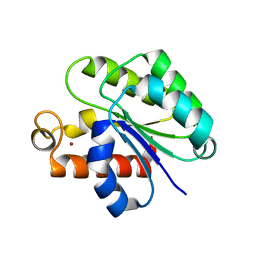 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain, Zn-bound | | 分子名称: | Acetyltransferase, ZINC ION | | 著者 | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | 登録日 | 2020-04-22 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6XCF
 
 | |
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
5NPP
 
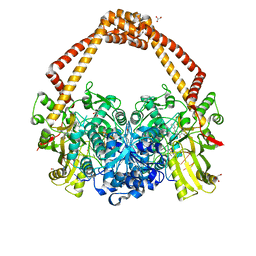 | | 2.22A STRUCTURE OF THIOPHENE2 AND GSK945237 WITH S.AUREUS DNA GYRASE AND DNA | | 分子名称: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DIMETHYL SULFOXIDE, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Bax, B.D, Chan, P.F, Stavenger, R.A. | | 登録日 | 2017-04-18 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
