9CJ0
 
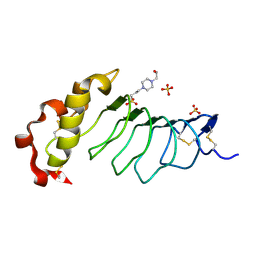 | |
9ILB
 
 | | HUMAN INTERLEUKIN-1 BETA | | 分子名称: | PROTEIN (HUMAN INTERLEUKIN-1 BETA) | | 著者 | Yu, B, Blaber, M, Gronenborn, A.M, Clore, G.M, Caspar, D.L.D. | | 登録日 | 1998-10-22 | | 公開日 | 1999-01-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
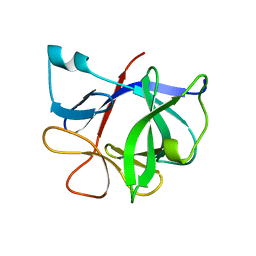
8ZH8
 
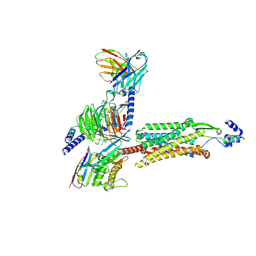 | | Human GPR103 -Gq complex bound to QRFP26 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Iwama, A, Akasaka, H, Sano, F.K, Oshima, H.S, Shihoya, W, Nureki, O. | | 登録日 | 2024-05-10 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structure and dynamics of the pyroglutamylated RF-amide peptide QRFP receptor GPR103.
Nat Commun, 15, 2024
|
|
9CWS
 
 | |
9BDD
 
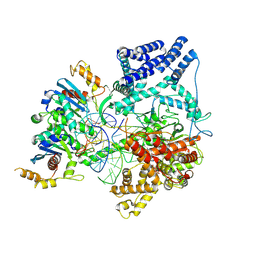 | | Cryo-EM Structure of Non-Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | | 分子名称: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase, mitochondrial, ... | | 著者 | Herbine, K.H, Nayak, A.R, Temiakov, D. | | 登録日 | 2024-04-11 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
9EO2
 
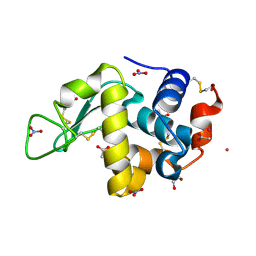 | |
9EM2
 
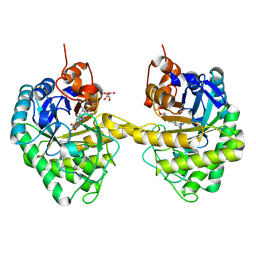 | |
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.423 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9EUO
 
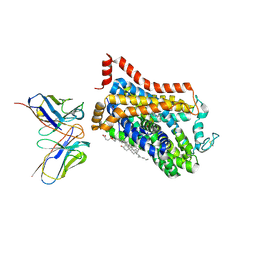 | | Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor | | 分子名称: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | 著者 | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | 登録日 | 2024-03-27 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9ETU
 
 | |
9EUP
 
 | | Inhibitor-free outward-open structure of Drosophila dopamine transporter | | 分子名称: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | 著者 | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | 登録日 | 2024-03-27 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 2024
|
|
9CV0
 
 | |
8U55
 
 | |
9FMN
 
 | | Structure of Human PADI6 | | 分子名称: | Protein-arginine deiminase type-6 | | 著者 | Mouilleron, S, Walport, L, Williams, J, Marsh, A.J, Hernandez Trapero, R. | | 登録日 | 2024-06-06 | | 公開日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural insight into the function of human peptidyl arginine deiminase 6.
Comput Struct Biotechnol J, 23, 2024
|
|
4W66
 
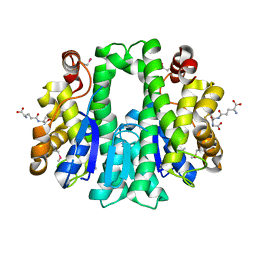 | |
7OW7
 
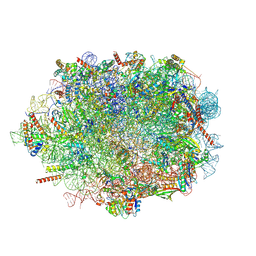 | |
4W6X
 
 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF7 | | 分子名称: | F18 fimbrial adhesin AC, Nanobody NbFedF7 | | 著者 | Moonens, K, De Kerpel, M, Coddens, A, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | 登録日 | 2014-08-21 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
7P2E
 
 | | Human mitochondrial ribosome small subunit in complex with IF3, GMPPMP and streptomycin | | 分子名称: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | 著者 | Itoh, Y, Khawaja, A, Singh, V, Rorbach, J, Amunts, A. | | 登録日 | 2021-07-05 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structure of the mitoribosomal small subunit with streptomycin reveals Fe-S clusters and physiological molecules.
Elife, 11, 2022
|
|
8TE7
 
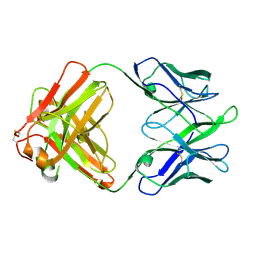 | | Structure of TRNM-f.01 | | 分子名称: | TRNM-f.01 Fab Heavy Chain, TRNM-f.01 Fab Light Chain | | 著者 | Bender, M.F, Olia, A.S, Kwong, P.D. | | 登録日 | 2023-07-05 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Broad and Potent HIV-1 Neutralization in Fusion Peptide-primed SHIV-boosted Macaques
To Be Published
|
|
8U1I
 
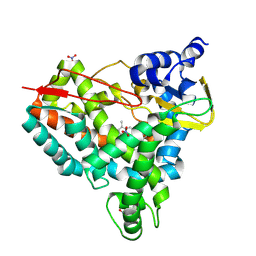 | | Crystal structure of SyoA bound to 4-methylsyringol | | 分子名称: | 2,6-dimethoxy-4-methylphenol, Cytochrome P450, NITRATE ION, ... | | 著者 | Harlington, A.C, Shearwin, K.E, Bell, S.G, Whelan, F. | | 登録日 | 2023-09-01 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structural insights into the S-lignin O-demethylase SyoA
To Be Published
|
|
8U09
 
 | | Crystal structure of substrate-free SyoA | | 分子名称: | ACETATE ION, Cytochrome P450, GLYCEROL, ... | | 著者 | Harlington, A.H, Shearwin, K.E, Bell, S.G, Whelan, F. | | 登録日 | 2023-08-28 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural insights into the S-lignin O-demethylase SyoA
To Be Published
|
|
8U19
 
 | | Crystal structure of SyoA bound to syringol | | 分子名称: | 2,6-dimethoxyphenol, Cytochrome P450, MAGNESIUM ION, ... | | 著者 | Harlington, A.C, Shearwin, K.E, Bell, S.G, Whelan, F. | | 登録日 | 2023-08-31 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Structural insights into the S-lignin O-demethylase SyoA
To Be Published
|
|
8TGZ
 
 | | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | 著者 | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | 登録日 | 2023-07-13 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
